Optimizing CRISPR Gene Editing Efficiency: From AI Design to Clinical Validation
This article provides a comprehensive guide for researchers and drug development professionals on optimizing CRISPR gene editing efficiency.
Optimizing CRISPR Gene Editing Efficiency: From AI Design to Clinical Validation
Abstract
This article provides a comprehensive guide for researchers and drug development professionals on optimizing CRISPR gene editing efficiency. It explores foundational AI-driven editor design, advanced methodological applications for therapeutic development, practical troubleshooting for common experimental challenges, and rigorous validation frameworks for assessing editing outcomes. Covering the latest advancements from 2025 research, the content synthesizes cutting-edge strategies including AI-generated editors like OpenCRISPR-1, novel delivery systems such as LNPs and VLPs, and cell-type specific optimization approaches to enhance both research and clinical applications.
The Next Generation: How AI and Novel CRISPR Systems Are Redefining Editing Efficiency
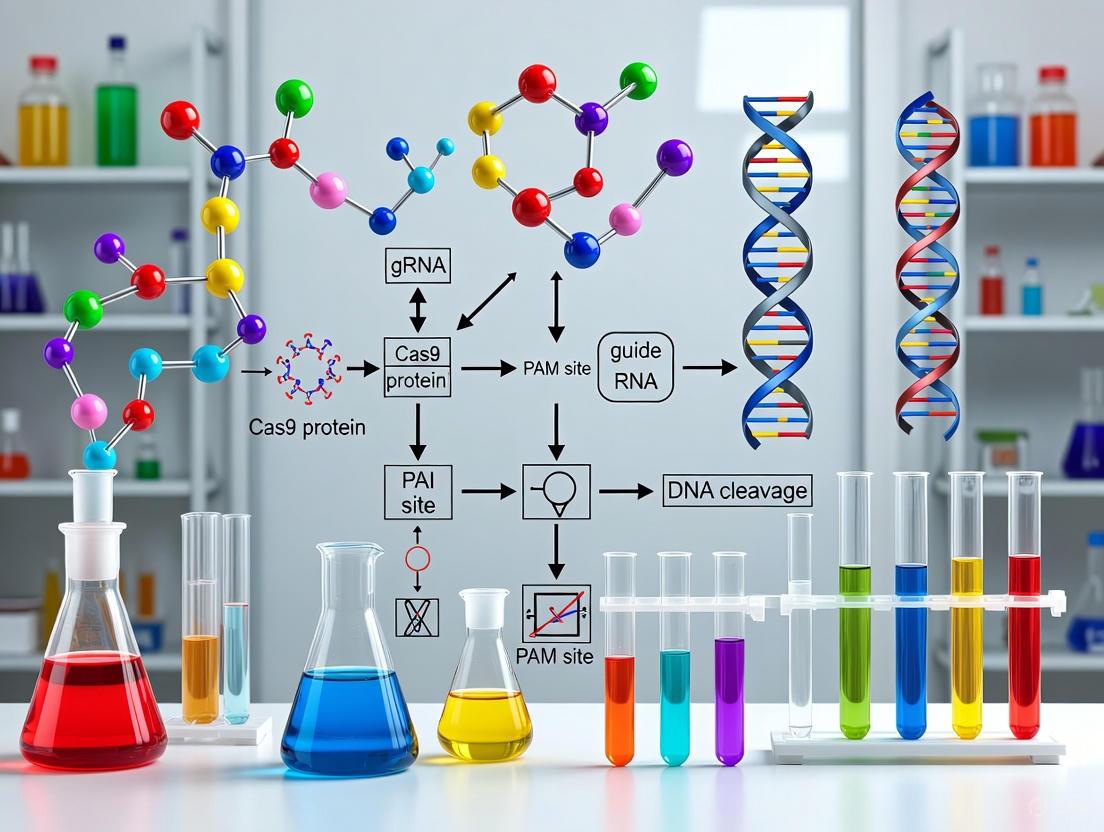
The integration of large language models (LLMs) into protein design represents a paradigm shift in how researchers approach CRISPR gene editing optimization. By treating protein sequences as a specialized "language" with its own grammatical rules and semantic relationships, these AI systems can generate novel protein editors with enhanced properties. This technical support center addresses the practical challenges researchers face when implementing these cutting-edge tools in their CRISPR efficiency optimization work. The following guides and resources will help you troubleshoot common issues, understand key methodologies, and leverage the latest AI-powered platforms to advance your gene editing research.
Frequently Asked Questions (FAQs): AI-Driven Protein Design
FAQ 1: How can a language model possibly understand or design a protein? Proteins can be treated as a "language" where the 20 amino acids constitute an alphabet. Motifs and domains are analogous to words and sentences, with the entire sequence encoding structure and function, much like a sentence conveys meaning. LLMs trained on vast protein databases learn the complex "grammar" and "syntax" that dictate stable, functional protein folds, allowing them to generate novel, valid sequences [1].
FAQ 2: What are the key advantages of using AI like ProtET over traditional protein engineering methods? Traditional methods are often slow, labor-intensive, and rely on single-task models. AI platforms like ProtET introduce a flexible, controllable approach using simple text instructions (e.g., "increase thermostability"), allowing researchers to explicitly guide protein editing for specific functions without being limited to a single predefined task [2]. This dramatically accelerates the exploration of the vast combinatorial space of potential protein edits.
FAQ 3: My AI-designed Cas protein has low editing efficiency in primary human cells. What could be the issue? Editing efficiency in therapeutically relevant primary cells (like lymphocytes) is highly dependent on nuclear delivery. A key strategy is to optimize nuclear localization signals (NLS). Recent studies show that incorporating hairpin internal NLS (hiNLS) sequences within the Cas9 backbone, rather than using terminally fused NLS, enhances nuclear import and can significantly boost editing efficiency in human primary T cells [3].
FAQ 4: How can I use AI to reduce the off-target effects of my CRISPR system? Off-target effects are often linked to the Cas enzyme's recognition of protospacer adjacent motifs (PAMs). Machine learning platforms like PAMmla can predict the properties of millions of CRISPR-Cas9 enzymes, enabling the selection or design of variants with more precise PAM recognition. This customizability minimizes off-target editing and expands the range of targetable genomic sites [4].
FAQ 5: What does "text-guided design" mean in the context of a tool like ProtET? Text-guided design allows a researcher to provide natural language instructions to the AI model to direct protein edits. For example, you can input a command like "optimize the heavy chain complementarity-determining region (CDR) for increased antigen binding affinity." The model, trained on millions of protein-text pairs, interprets this instruction and generates the corresponding protein sequence modifications [2].
Troubleshooting Common Experimental Issues
Issue 1: Low Knock-In Efficiency in Primary B Cells
Problem: CRISPR/Cas9-mediated knock-in efficiency is unacceptably low in primary human B cells, which are often quiescent and favor the error-prone NHEJ repair pathway over HDR [5].
Solutions:
- Optimize HDR Template Design: For short insertions (e.g., tags or point mutations), use single-stranded DNA donors with 30-60 nt homology arms. For larger insertions (e.g., fluorescent proteins), use double-stranded plasmid templates with 200-300 nt homology arms [5].
- Consider Strand Preference: If the edit is more than 5-10 bp from the Cas9 cut site, design your template for the targeting strand for PAM-proximal edits, and the non-targeting strand for PAM-distal edits [5].
- Utilize AI Design Tools: Use platforms like CRISPR-GPT to analyze your specific experimental goals, suggest optimal sgRNA targets, and predict potential off-target effects that could compromise efficiency [6].
Issue 2: Poor Stability or Expression of AI-Designed Protein
Problem: A novel protein editor generated by an AI model (e.g., a custom Cas variant) shows poor soluble expression or low stability in vivo.
Solutions:
- Employ Text-Guided Stabilization: Use a model like ProtET with text instructions focused on stability, such as "improve thermal stability" or "increase soluble expression yield." The model can then propose stabilizing mutations [2].
- Leverage Pre-Trained Models: Platforms like PAMmla are trained on scalable protein engineering data and can predict functional enzymes from a space of 64 million variants, helping you select a stable, well-behaved candidate from the outset [4].
Issue 3: High Off-Target Activity with Novel Editors
Problem: A newly designed nuclease shows high on-target activity but also unacceptably high levels of off-target editing.
Solutions:
- Select High-Fidelity Variants: Use machine learning-predicted Cas9 enzymes that are specifically tuned for enhanced precision. The PAMmla platform, for instance, can identify variants with stricter PAM recognition, a key factor in reducing off-target effects [4].
- Validate with AI Analysis: Before experimental validation, use tools like CRISPR-GPT to analyze your sgRNA design and predict the likelihood and location of potential off-target edits based on historical data [6].
Performance Data of AI Platforms for Protein Design
Table 1: Comparison of Key AI Platforms for Protein and CRISPR Editor Design
| Platform Name | Primary Function | Key Input | Reported Outcome/Advantage | Source/Reference |
|---|---|---|---|---|
| ProtET | Controllable protein editing | Natural language instructions (e.g., "increase stability") | 16.67-16.90% improvement in stability; optimized antibody binding | [2] |
| PAMmla | Cas9 enzyme property prediction | Scalable sequence data | Predicts properties of 64 million Cas9 variants for precise editing | [4] |
| CRISPR-GPT | Gene-editing experiment planning | Experimental goals & gene sequences | Automates experimental design; enables successful first-attempt edits by novices | [6] |
| hiNLS Cas9 | Enhanced nuclear delivery | Cas9 sequence with internal NLS motifs | Improved editing efficiency in primary human T cells | [3] |
Experimental Protocols for Key Methodologies
Protocol 1: Implementing hiNLS Cas9 for Enhanced Editing in Primary Cells
This protocol is adapted from strategies shown to enhance CRISPR editing efficiency in therapeutically relevant primary human lymphocytes [3].
- Vector Construction: Engineer your Cas9 expression vector to incorporate hairpin internal nuclear localization signal (hiNLS) sequences at selected sites within the Cas9 backbone, as opposed to the traditional terminally-fused NLS.
- Protein Production: Express and purify the hiNLS-Cas9 protein. Studies indicate these constructs can be produced with high purity and yield, supporting manufacturing scalability.
- Ribonucleoprotein (RNP) Complex Formation: Complex the purified hiNLS-Cas9 protein with your target-specific sgRNA to form RNP complexes.
- Cell Delivery: Deliver the RNP complexes into primary human T cells or other target cells via electroporation. The hiNLS design facilitates rapid nuclear import, which is critical for high efficiency when using transient RNP delivery.
- Validation: Assess editing efficiency at the target locus (e.g., B2M or TRAC) using next-generation sequencing or T7E1 assay. Compare against a terminal-NLS control.
Protocol 2: Using Text-Guided AI for Protein Optimization
This protocol outlines the workflow for using a tool like ProtET to optimize a protein of interest [2].
- Define Goal: Formulate a clear, text-based instruction that defines the desired functional outcome. Example: "Enhance the catalytic activity of [Your Enzyme] for substrate [X]."
- Input Sequence: Provide the AI model (ProtET) with the wild-type amino acid sequence of your target protein.
- Generate Variants: Run the model with your text instruction to receive a set of proposed protein sequence variants.
- Synthesize and Clone: Synthesize the genes encoding the top AI-predicted variants and clone them into an appropriate expression vector.
- Experimental Testing: Express and purify the variant proteins. Test them in relevant functional assays (e.g., enzyme activity assays, thermal shift assays for stability, or SPR for binding affinity).
Workflow and Pathway Visualizations
AI-Driven Protein Editor Design Workflow
Troubleshooting Low Knock-In Efficiency
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Research Reagents for AI-Powered CRISPR Editor Development
| Reagent / Material | Function / Description | Example Use Case |
|---|---|---|
| hiNLS-Cas9 Constructs | Cas9 variants with internal nuclear localization signals for enhanced nuclear import and editing efficiency in primary cells. | Boosting knock-in efficiency in hard-to-transfect primary human T or B cells for immunotherapy research [3]. |
| Machine Learning-Predicted Cas Variants | Novel Cas enzymes (e.g., from PAMmla) designed for reduced off-target effects or altered PAM recognition. | Performing highly precise edits in genotherapeutic applications where safety is paramount [4]. |
| Single-Stranded DNA (ssDNA) HDR Donor | Short, single-stranded DNA oligonucleotides with 30-60 nt homology arms, used as a repair template for introducing small edits. | Introducing point mutations or small tags (e.g., FLAG, HIS) into a specific genomic locus [5]. |
| Plasmid HDR Donor Templates | Double-stranded DNA plasmids containing the insert (e.g., fluorescent protein) flanked by long homology arms (200-300 nt). | Inserting larger genetic elements, such as reporter genes or degron tags, into the genome [5]. |
| AI-Generated Protein Variants | Novel protein sequences generated by platforms like ProtET, optimized for specific properties like stability or binding. | Rapidly engineering improved enzymes, therapeutic antibodies, or stabilized CRISPR editors without extensive manual screening [2]. |
| CRISPR-GPT Analysis Report | An AI-generated plan suggesting sgRNA designs, potential pitfalls, and troubleshooting advice for a gene-editing experiment. | Guiding novice researchers or optimizing complex experimental designs to save time and resources [6]. |
FAQs: Novel Cas Proteins and Systems
Q1: Why is the discovery of novel Cas proteins important for therapeutic development? The discovery of novel Cas proteins is crucial because different Cas variants have unique properties that can overcome limitations of the commonly used SpCas9. For instance, some newly identified Cas proteins are smaller in size. This smaller size is vital for packaging the entire CRISPR system into delivery vehicles with limited capacity, such as Adeno-Associated Viruses (AAVs), which are commonly used for in vivo gene therapy. Furthermore, novel Cas proteins often recognize different Protospacer Adjacent Motif (PAM) sequences, expanding the range of DNA sites that can be targeted within the genome for editing [7] [8].
Q2: What are the main challenges when working with a newly discovered Cas protein in the lab? The primary challenges involve characterization and optimization. Researchers must first thoroughly characterize the new protein's properties, including its specific PAM requirement, on-target editing efficiency, and potential for off-target effects. Subsequently, the entire system—including the guide RNA and delivery method—often needs to be re-optimized for the new nuclease to function effectively in the target cell type. This process can be time-consuming and requires careful experimental validation [9] [10].
Q3: How do base editors and prime editors differ from traditional Cas nucleases? Traditional Cas nucleases, like Cas9, create double-strand breaks (DSBs) in DNA, which rely on the cell's repair mechanisms and can lead to unpredictable insertions or deletions (indels). In contrast, base editors use a catalytically impaired Cas nuclease fused to a deaminase enzyme to directly convert one base into another (e.g., C to T or A to G) without creating a DSB. Prime editors are even more versatile, using a Cas9 nickase fused to a reverse transcriptase to directly write new genetic information into a target DNA site based on a prime editing guide RNA (pegRNA) template, also without requiring a DSB. These systems offer greater precision and reduce unwanted editing byproducts [10] [11].
Q4: What delivery methods are most promising for CRISPR therapies using novel systems? Delivery remains a central challenge. While viral vectors like AAVs are efficient, they can trigger immune responses and have limited packaging capacity. Non-viral methods are increasingly promising:
- Lipid Nanoparticles (LNPs): Effectively deliver CRISPR components as RNA or ribonucleoprotein (RNP) complexes. They are particularly good for targeting liver cells and allow for potential re-dosing, as seen in recent clinical trials [7] [12] [13].
- Electroporation: Highly effective for ex vivo editing of hard-to-transfect cells like primary T cells and hematopoietic stem cells [7].
- Novel Nanostructures: Emerging systems, like lipid nanoparticle spherical nucleic acids (LNP-SNAs), have shown in lab studies to boost cell uptake and gene-editing efficiency while reducing toxicity compared to standard LNPs [13].
Troubleshooting Guides
Table 1: Troubleshooting Common Issues with Novel Cas Proteins
| Problem | Possible Cause | Solution |
|---|---|---|
| Low Editing Efficiency | Non-optimal guide RNA (gRNA) design for the new Cas protein. | Redesign gRNAs using tools specific to the Cas variant, ensuring target specificity and avoiding secondary structures [9]. |
| Inefficient delivery of CRISPR components into target cells. | Optimize delivery method (e.g., electroporation parameters, LNP formulation). Use a reporter system to confirm successful delivery [7] [9]. | |
| Low expression or activity of the novel Cas protein in the host cell. | Verify protein expression with a Western blot. Use a codon-optimized sequence for your host organism and confirm promoter compatibility [9]. | |
| High Off-Target Effects | The novel Cas protein may have lower intrinsic fidelity. | Utilize computational tools to predict and scan for potential off-target sites. Consider using high-fidelity engineered versions of the nuclease if available [9] [10]. |
| gRNA sequence has high similarity to other genomic regions. | Design gRNAs with maximal on-target and minimal off-target homology. Perform deep sequencing to comprehensively assess off-target activity [14] [9]. | |
| High Cell Toxicity | Overexpression of the CRISPR components is stressful to cells. | Titrate the amount of Cas/gRNA delivered; use lower, more effective doses. For novel Cas proteins, delivering pre-assembled RNP complexes can reduce the duration of nuclease activity and lower toxicity [7] [9]. |
| The delivery method itself (e.g., electroporation) is causing stress. | Optimize delivery parameters to improve cell viability post-transfection. For electroporation, adjust voltage and pulse length [7]. | |
| Failure to Knock-in Gene | The DNA repair pathway (HDR) is inefficient in your cell type. | Use HDR-enhancing reagents and deliver CRISPR during the S/G2 phase of the cell cycle (e.g., by cell cycle synchronization). For large insertions, consider using advanced systems like retron-based editing [7] [15]. |
| The donor DNA template is not co-localizing with the cut site. | Use methods that facilitate co-delivery, such as electroporation of RNP with a single-stranded DNA (ssODN) donor or the use of virus-like particles (VLPs) [7] [8]. |
Table 2: Advanced Troubleshooting: Scaling Up and Validation
| Challenge | Consideration & Strategy |
|---|---|
| Multiplexed Editing | Delivering multiple gRNAs simultaneously increases payload size and complexity. Lentiviral vectors or electroporation are suitable methods. Carefully assess potential synergistic toxicity or off-target effects when targeting multiple loci [7]. |
| In Vivo Validation | Efficient delivery to the target tissue is the major hurdle. Select delivery vectors (e.g., LNPs, AAVs) with known tropism for your organ of interest. New systems like LNP-SNAs aim to improve tissue targeting [13]. Always include controls to distinguish between editing efficiency and delivery efficiency. |
| Detection of Precise Edits | Standard genotyping methods may not detect precise base changes or small insertions. Use a combination of methods: T7E1 or Surveyor assays for initial cleavage detection, followed by Sanger sequencing or next-generation sequencing (NGS) for precise characterization of the edits [9]. |
Experimental Protocols
Protocol 1: Initial Characterization of a Novel Cas Protein's PAM Requirement
Objective: To empirically determine the Protospacer Adjacent Motif (PAM) sequence essential for the novel Cas nuclease to recognize and cleave DNA.
Materials:
- Plasmid library containing a randomized PAM sequence adjacent to a constant target site.
- The novel Cas nuclease and its associated guide RNA (gRNA) expression constructs.
- A suitable host cell line (e.g., HEK293T for high transfection efficiency).
- Next-Generation Sequencing (NGS) capabilities.
Methodology:
- Library Delivery: Co-transfect the PAM library and the CRISPR constructs (Cas + gRNA) into the host cells.
- Selection Pressure: After 48-72 hours, harvest the plasmid DNA from the cells. The functional cleavage by the Cas nuclease will destroy plasmids with non-permissive PAMs, enriching for plasmids with functional PAM sequences.
- Sequencing and Analysis: Amplify the region containing the randomized PAM from the harvested plasmid pool and subject it to NGS. Compare the sequence reads before and after selection to identify the PAM sequences that have been enriched, revealing the required PAM motif [10].
Protocol 2: Assessing Editing Efficiency and Specificity
Objective: To quantify the on-target efficiency and profile the off-target activity of a novel CRISPR system.
Materials:
- Target cells.
- Validated gRNA for your target locus.
- Optimized method for delivering the novel CRISPR system (e.g., RNP via electroporation).
- T7 Endonuclease I or Surveyor Mutation Detection Kit.
- PCR reagents and NGS platform.
Methodology:
- Delivery and Culture: Deliver the CRISPR components to the target cells and culture for several days to allow editing and repair.
- Genomic DNA Extraction: Harvest genomic DNA from the edited cell population.
- On-Target Efficiency Analysis:
- PCR: Amplify the genomic region surrounding the target site.
- T7E1/Surveyor Assay: Digest the PCR amplicon with the mismatch-sensitive enzyme. Cleaved bands on a gel indicate successful editing. The intensity of the bands can be used for a rough efficiency estimate.
- NGS (Gold Standard): Amplify the target region and perform NGS. This provides a quantitative measure of the percentage of alleles containing insertions, deletions, or precise edits [9].
- Off-Target Analysis:
Research Workflow and Pathways
Diagram 1: Novel Cas Protein Characterization Workflow
Diagram 2: CRISPR-Cas Genome Editing and Repair Pathways
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Reagents for Working with Novel Cas Systems
| Reagent / Material | Function & Application |
|---|---|
| Codon-Optimized Cas Expression Vector | Ensures high-level expression of the novel Cas protein in the target host organism (e.g., human cells). |
| Guide RNA (gRNA) Expression Constructs | Delivers the targeting RNA component; can be on a separate plasmid or in an all-in-one vector with the Cas gene [7]. |
| Synthetic sgRNA or crRNA/tracrRNA | Chemically synthesized guide RNAs offer high purity and reduced immune activation in therapeutic contexts; can be complexed with Cas protein to form RNP complexes [7] [13]. |
| Lipid Nanoparticles (LNPs) | A non-viral delivery vehicle for in vivo delivery of CRISPR mRNA, sgRNA, or donor templates. Particularly effective for liver targets [12] [13]. |
| Electroporation System | A physical delivery method highly effective for ex vivo editing of hard-to-transfect primary cells (e.g., T cells, stem cells) with RNPs [7]. |
| NGS Mutation Detection Kit | Provides the reagents for preparing sequencing libraries to accurately quantify on-target and off-target editing frequencies. |
| HDR Donor Template (ssODN/dsDNA) | Single-stranded oligodeoxynucleotides (ssODNs) for small edits; double-stranded DNA (dsDNA) for larger insertions. Critical for precise gene correction or knock-in [7] [15]. |
| Cell Type-Specific Culture Media | Essential for maintaining the viability and function of sensitive primary cells during and after the CRISPR editing process. |
The integration of artificial intelligence (AI) with CRISPR gene editing represents a transformative advancement in biotechnology, enabling researchers to move beyond naturally derived systems to computationally designed editors with enhanced properties. AlphaFold, DeepMind's revolutionary protein structure prediction network, has emerged as a pivotal tool in this paradigm shift. By accurately predicting three-dimensional protein structures from amino acid sequences, AlphaFold provides critical structural insights that accelerate the rational optimization of gene editing tools [16] [17]. This technical support resource examines how structural biology powered by AlphaFold is addressing fundamental challenges in CRISPR editor efficiency, specificity, and functionality, providing researchers with practical methodologies to enhance their experimental outcomes.
The optimization of CRISPR systems has traditionally relied on iterative experimental screening—a time-consuming and often unpredictable process. AlphaFold circumvents these limitations by enabling computational structural analysis of CRISPR complexes before experimental validation. Research demonstrates that AlphaFold predictions achieve accuracy "competitive with experimental structures in a majority of cases," providing reliable structural models for rational design [16]. This capability is particularly valuable for characterizing CRISPR-Cas proteins with unresolved experimental structures, such as PspCas13b, where AlphaFold predictions with high per-residue confidence scores (pLDDT) have enabled the identification of functional domains and strategic splitting sites for engineered editor systems [18].
Frequently Asked Questions (FAQs)
Q1: How can AlphaFold improve gRNA design for CRISPR systems? AlphaFold structural predictions enable rational guide RNA optimization by modeling the molecular interactions between Cas proteins and their RNA guides. For instance, with the compact CRISPR/Cas12f1 system, AlphaFold 3 simulations revealed suboptimal intramolecular pairing within the tracrRNA component that disrupted its interaction with crRNA. Researchers used these structural insights to strategically truncate the 3' end of tracrRNA, eliminating disruptive pairing and significantly enhancing trans-cleavage activity. Subsequent design of a single guide RNA (sgRNA) further improved system performance, demonstrating how AlphaFold-driven structural analysis can optimize nucleic acid components for enhanced editor function [19].
Q2: Can AlphaFold predict structures for novel, AI-designed CRISPR proteins? Yes, AlphaFold can reliably fold artificially generated CRISPR proteins that diverge significantly from natural sequences. In a landmark study, researchers used large language models to generate millions of novel CRISPR-Cas sequences, with generated Cas9-like proteins averaging only 56.8% sequence identity to any natural counterpart. Despite this divergence, AlphaFold2 confidently predicted structures for 81.65% of these AI-generated proteins (mean pLDDT > 80), enabling computational validation of structural integrity before experimental testing. This capability provides a crucial bridge between AI-based protein generation and functional implementation [20].
Q3: How accurate are AlphaFold predictions for CRISPR protein engineering? Validation studies demonstrate exceptional accuracy for AlphaFold in predicting structures of CRISPR-related proteins. In comprehensive assessments against experimentally determined structures, AlphaFold achieved median backbone accuracy of 0.96 Å RMSD95 (approximately the width of a carbon atom) [16]. For anti-CRISPR proteins—small proteins used to inhibit CRISPR-Cas systems—AlphaFold2 predictions showed no significant difference in accuracy compared to its performance in the CASP14 assessment, with TM-scores indicating highly reliable structural models [21]. This precision enables confident use of predictions for rational engineering decisions.
Q4: What are the limitations of using AlphaFold for editor optimization? While transformative, AlphaFold has specific limitations. It may not fully capture conformational changes induced by crRNA binding or target DNA/RNA recognition [18]. Additionally, although AlphaFold can be inverted for de novo protein design, initial designs often require optimization of surface properties, as early trials produced designs with hydrophobic surface patches uncharacteristic of natural proteins [22]. For optimal results, researchers should complement AlphaFold predictions with experimental validation and molecular dynamics simulations where conformational flexibility is functionally important.
Troubleshooting Common Experimental Challenges
Challenge: Low Editing Efficiency with Novel Cas Variants
Problem: AI-designed Cas proteins exhibit poor editing activity in cellular environments despite proper expression.
Solution: Utilize AlphaFold to analyze structural completeness and active site conservation.
Step-by-Step Protocol:
- Generate structural models of your novel Cas variant using AlphaFold (via ColabFold or local installation) [16] [17].
- Compare the predicted structure to well-characterized natural Cas proteins (e.g., SpCas9), focusing on:
- Conservation of catalytic residues (RuvC, HNH for Cas9)
- Structural integrity of guide RNA binding regions
- Proper formation of protospacer adjacent motif (PAM) interaction sites
- Identify structural deviations that might impair function, such as:
- Misfolded catalytic domains
- Obstructed substrate binding channels
- Incomplete formation of key structural motifs
- Implement structure-guided repairs by reverting non-functional mutations back to conserved residues while maintaining novel beneficial substitutions.
- Validate computationally by re-predicting the structure of repaired variants before experimental testing.
Table: Critical Functional Domains to Verify in AI-Designed Cas Proteins
| CRISPR System | Essential Functional Domains | Key Structural Features to Verify |
|---|---|---|
| Cas9-like | RuvC, HNH, REC lobe, PAM-interacting | Catalytic residue geometry, DNA/RNA binding cleft formation |
| Cas12-like | RuvC, OBD, Nuc | Electrostatic surface for DNA engagement, cleavage site accessibility |
| Cas13-like | HEPN domains, guide RNA binding region | Conserved catalytic arginines, nucleotide binding pockets |
Challenge: Engineering Inducible CRISPR Systems
Problem: Achieving precise temporal control over CRISPR activity through split protein systems.
Solution: Use AlphaFold to identify optimal split sites that minimize background activity while maintaining inducibility.
Step-by-Step Protocol (Based on paCas13 Development):
- Obtain a high-confidence structural model of your target Cas protein using AlphaFold with multiple recycling steps (3-5 cycles) to refine model quality [18].
- Identify candidate split sites in solvent-accessible loop regions using these criteria:
- Located away from catalytic centers and substrate binding interfaces
- Positioned in flexible linkers between structured domains
- Avoid terminal HEPN domains for Cas13 systems
- Generate structural models of split fragments to assess:
- Residual interface compatibility using MaSIF-site or similar tools
- Electrostatic complementarity of fragment surfaces
- Potential for spontaneous reconstitution (high in split sites N550/C551, N624/C625 for PspCas13b)
- Select split sites with moderate interface areas that require dimerization domains for stable association (e.g., N351/C352 for PspCas13b).
- Validate fragment structures individually and in complex with dimerization domains to ensure proper folding when reconstituted.
Diagram: Workflow for Identifying Optimal Split Sites in Cas Proteins
Challenge: Optimizing Base Editor Efficiency
Problem: Low editing efficiency in base editor systems due to suboptimal spatial arrangement of catalytic domains.
Solution: Use AlphaFold to model the spatial relationship between deactivated Cas domains and effector proteins.
Step-by-Step Protocol:
- Construct a structural model of your base editor complex, including:
- Deactivated Cas protein (dCas9, dCas13, etc.)
- Tethered effector domain (deaminase, acetyltransferase, etc.)
- Appropriate linker sequences between domains
- Analyze the spatial orientation of the catalytic site relative to the target nucleotide:
- Measure distances between catalytic residues and target base
- Identify potential steric clashes with Cas protein backbone
- Assess flexibility and accessibility of the target region
- Systematically vary linker length and composition, re-predicting structures for each variant.
- Select configurations that position catalytic residues within optimal range of the target base (typically 5-15Å).
- Validate top candidates through molecular dynamics simulations if possible, then experimental testing.
Research Reagent Solutions
Table: Essential Computational and Experimental Resources for AlphaFold-Guided Editor Optimization
| Resource Category | Specific Tools/Solutions | Application in Editor Optimization |
|---|---|---|
| Structure Prediction | AlphaFold2, AlphaFold 3, ColabFold | Protein-RNA complex modeling, guide RNA optimization, split system design [19] [16] |
| Structure Analysis | MaSIF-site, PyMOL, ChimeraX | Interface analysis, electrostatic potential mapping, conformational assessment [18] |
| Sequence Generation | ProGen2, CRISPR-Cas Atlas | Generating novel Cas variants with expanded diversity [20] |
| Validation Metrics | pLDDT, predicted TM-score, RMSD | Assessing prediction reliability, model quality assurance [16] [20] |
| Experimental Testing | Dual-luciferase assays, mammalian cell editing screens | Functional validation of designed editors [18] |
Advanced Applications and Methodologies
De Novo Design of CRISPR Proteins
Beyond optimizing natural systems, AlphaFold enables the computational design of entirely novel CRISPR proteins. By inverting the AlphaFold network, researchers can generate sequences that fold into desired structures, though this approach may require optimization of surface properties [22]. The integration of large language models trained on CRISPR protein diversity has dramatically expanded this capability, generating Cas9-like effectors with 10.3-fold increased phylogenetic diversity compared to natural sequences [20].
Protocol for Assessing Novel CRISPR Protein Folds:
- Generate novel protein sequences using CRISPR-specific language models (e.g., ProGen2 fine-tuned on CRISPR-Cas Atlas) [20].
- Predict structures of generated sequences using AlphaFold2.
- Evaluate structural quality using confidence metrics (pLDDT, predicted TM-score).
- Verify conservation of key functional motifs despite sequence divergence.
- Select candidates with both high confidence scores and novel structural features for experimental characterization.
Leveraging Natural Anti-CRISPR Mechanisms
AlphaFold structural predictions have revealed remarkable diversity in anti-CRISPR proteins, providing insights into inhibition mechanisms that can inform editor optimization [21]. These natural inhibitory proteins employ diverse strategies, including direct Cas protein binding, active site occlusion, and even enzymatic modification of Cas complexes.
Protocol for Analyzing Anti-CRISPR Mechanisms:
- Acquire anti-CRISPR datasets from viral and prokaryotic genomes [21].
- Predict 3D structures using AlphaFold2 with high confidence (plDDT > 70 recommended).
- Classify inhibition mechanisms through structural analysis:
- Competitive binding to DNA/RNA interaction sites
- Allosteric inhibition through conformational stabilization
- Enzymatic inactivation (e.g., acetylation of active sites)
- Apply insights to engineer regulatable Cas systems or enhance specificity through inhibitory domain incorporation.
Diagram: From Anti-CRISPR Mechanisms to Editor Applications
Quantitative Assessment Framework
Table: Key Metrics for Evaluating AlphaFold-Guided Editor Optimization
| Performance Dimension | Evaluation Metrics | Benchmark Values |
|---|---|---|
| Prediction Accuracy | TM-score, RMSD, plDDT | TM-score > 0.8 (high confidence), Backbone accuracy ~0.96Å [16] |
| Editor Efficiency | Editing rate, Specificity index | Comparable or improved vs. SpCas9 baseline [20] |
| Novelty | Sequence identity to natural proteins, Phylogenetic diversity | 40-60% identity to nearest natural protein [20] |
| Functional Success | Experimental success rate, Melting temperature | 7/39 de novo designs folded with high Tm [22] |
| Process Acceleration | Design-to-validation timeline | Weeks vs. months for traditional engineering [23] |
This technical support resource demonstrates how AlphaFold-driven structural biology provides a powerful framework for addressing persistent challenges in CRISPR editor optimization. By integrating these computational methodologies into their research pipeline, scientists can accelerate the development of next-generation genome editing tools with enhanced precision, functionality, and therapeutic potential.
The discovery of the CRISPR-Cas9 system has revolutionized genetic engineering, with Streptococcus pyogenes Cas9 (SpCas9) serving as the foundational enzyme for countless applications. However, inherent limitations of wild-type SpCas9, including off-target effects, strict Protospacer Adjacent Motif (PAM) requirements, and occasional low editing efficiency, have driven the development of enhanced variants and orthologs. This technical resource center provides a comprehensive guide to these advanced tools, offering researchers detailed protocols, troubleshooting advice, and reagent solutions to optimize genome editing efficiency.
FAQs: Selecting High-Efficiency CRISPR Systems
1. What are the primary advantages of using high-fidelity Cas9 variants over wild-type SpCas9? High-fidelity Cas9 variants are engineered to drastically reduce off-target editing while maintaining robust on-target activity. Wild-type SpCas9 can tolerate mismatches between the guide RNA and target DNA, leading to unintended genomic modifications. Variants like eSpCas9(1.1), SpCas9-HF1, HypaCas9, and evoCas9 incorporate mutations that disrupt non-specific interactions with the DNA backbone or enhance the enzyme's proofreading capability, thereby increasing its discrimination against off-target sites [24]. For example, the Alt-R S.p. HiFi Cas9 nuclease dramatically reduces off-target effects compared to the wild-type enzyme [25].
2. Which Cas enzymes should I consider for targeting genomic regions lacking an NGG PAM sequence? The requirement for an NGG PAM sequence adjacent to the target site can be a significant limitation. Fortunately, several engineered Cas variants and natural orthologs with altered PAM compatibilities are now available:
- xCas9: Recognizes NG, GAA, and GAT PAMs, and also offers increased fidelity [24].
- SpCas9-NG: Engineered to recognize NG PAMs, increasing the number of potential target sites in the genome [24].
- SpG: A variant that recognizes NGN PAMs, offering broader targeting range [24].
- SpRY: A nearly PAM-less variant that recognizes NRN (preferring NGG) and NYN (preferning NGT) PAMs, providing exceptional flexibility [24].
- SaCas9: A Staphylococcus aureus-derived Cas9 that is smaller than SpCas9 and recognizes the NNGRRT PAM [25].
- CjCas9: A compact Cas9 from Campylobacter jejuni with a NNNNACAC PAM requirement [25].
3. Are there fully novel Cas proteins not found in nature? Yes, generative artificial intelligence is now being used to design novel CRISPR-Cas proteins. A prime example is OpenCRISPR-1, a Cas9-like protein designed by Profluent Bio using large language models. OpenCRISPR-1 is reported to have comparable on-target efficiency to SpCas9 while demonstrating a 95% reduction in off-target editing across multiple genomic sites. It is composed of 1,380 amino acids and contains 403 mutations compared to SpCas9, making it a truly AI-generated editor [26].
4. How does Cas12a differ from Cas9, and when should I use it? Cas12a (formerly known as Cpf1) is a distinct type of CRISPR system with several unique features that make it suitable for specific applications [27]:
- PAM Sequence: It recognizes a T-rich PAM (TTTV for Cas12a V3, TTTN for Cas12a Ultra), which is located 5' of the target site.
- Cleavage Mechanism: It creates staggered ends (offsets) in the target DNA, as opposed to the blunt ends created by Cas9. This can be beneficial for certain cloning strategies and may improve Homology-Directed Repair (HDR) efficiency.
- crRNA Processing: Cas12a processes its own guide RNA arrays, which can simplify multiplexing strategies where multiple genes are targeted simultaneously. The Alt-R Cas12a Ultra nuclease has higher on-target potency and a broader TTTN PAM recognition, increasing its target range [25].
Troubleshooting Guides
Problem: Low On-Target Editing Efficiency
Potential Causes and Solutions:
Cause 1: Suboptimal guide RNA (gRNA) design.
- Solution: Ensure your gRNA has sufficient GC content. A study in grapevine found that editing efficiency increased proportionally with sgRNA GC content, with 65% GC content yielding the highest efficiency [28]. Use established online tools to select a gRNA with high predicted efficiency and minimal off-targets. Also, verify that the gRNA is expressed correctly in your system.
Cause 2: Persistent non-selective DNA binding by the Cas nuclease.
- Solution: Recent research indicates that reduced PAM specificity can lead to the Cas enzyme becoming stuck on non-target DNA, failing to engage the correct target sequence. If using a PAM-flexible variant like SpRY, consider switching to a more specific enzyme like a high-fidelity SpCas9 for targets with standard NGG PAMs [29].
Cause 3: Low expression or activity of the Cas protein in your cell type.
- Solution: Optimize the codon usage of the Cas gene for your target organism. Use a strong, cell-type-specific promoter to drive Cas expression. Validate protein expression using western blotting. For difficult-to-transfect cells, enriching for transfected cells via antibiotic selection or FACS sorting can improve results [30].
Problem: High Off-Target Editing
Potential Causes and Solutions:
Cause 1: Use of wild-type SpCas9 with a gRNA that has high sequence homology elsewhere in the genome.
- Solution: The most direct solution is to switch to a high-fidelity Cas9 variant such as eSpCas9(1.1), SpCas9-HF1, or HypaCas9 [24]. These mutants are designed to be less tolerant of gRNA-DNA mismatches. Alternatively, use the Cas9 nickase (Cas9n) system, which requires two adjacent gRNAs to create a double-strand break, dramatically increasing specificity [24].
Cause 2: High, prolonged expression of Cas9 and gRNA.
- Solution: Utilize transient expression systems, such as delivering the CRISPR machinery as a ribonucleoprotein (RNP) complex. RNP delivery leads to rapid degradation and reduced off-target effects compared to plasmid-based expression [31]. Employ self-inactivating vectors or inducible promoters to limit the duration of Cas9 activity.
Cause 3: Inadequate assessment of off-target sites.
- Solution: Move beyond biased computational prediction. Employ genome-wide, unbiased off-target detection methods like GUIDE-seq or Digenome-seq to get a comprehensive profile of nuclease activity [31]. This provides a more accurate picture of specificity.
Comparative Data Tables
Table 1: Engineered SpCas9 Variants for Enhanced Specificity and Altered PAM Recognition
| Variant Name | Key Feature | Mechanism | PAM Sequence | Best Use Case |
|---|---|---|---|---|
| eSpCas9(1.1) [24] | High-fidelity | Weakened non-target strand binding to reduce off-target cleavage. | NGG | Experiments requiring maximal on-target accuracy. |
| SpCas9-HF1 [24] | High-fidelity | Disrupted interactions with DNA phosphate backbone. | NGG | General high-specificity applications. |
| HypaCas9 [24] | High-fidelity | Enhanced proofreading and discrimination between on- and off-targets. | NGG | Sensitive genetic screens and therapeutic development. |
| xCas9 [24] | PAM-flexible & High-fidelity | Mutations in multiple domains to alter PAM recognition. | NG, GAA, GAT | Targeting regions with limited NGG sites, needing higher fidelity. |
| SpCas9-NG [24] | PAM-flexible | Engineered PAM-interacting domain. | NG | Significantly expands the targetable genome space. |
| SpRY [24] | Near PAM-less | Greatly relaxed PAM requirement. | NRN > NYN | Targeting genomic regions with no canonical PAM sequences. |
Table 2: Diverse Cas Nucleases and Their Properties
| Cas Nuclease | Origin | Size (aa) | PAM Sequence (5'—3') | Key Characteristics |
|---|---|---|---|---|
| SpCas9 [25] | Streptococcus pyogenes | 1,368 | NGG | The gold standard; well-characterized but has size and PAM limitations. |
| SaCas9 [25] | Staphylococcus aureus | 1,053 | NNGRRT | Smaller size beneficial for viral vector packaging (e.g., AAV). |
| CjCas9 [25] | Campylobacter jejuni | ~984 | NNNNACAC | Very compact; useful for delivery where size is a constraint. |
| AsCas12a [25] | Acidaminococcus sp. | 1,307 | TTTN | Creates staggered cuts; self-processes crRNAs; T-rich PAM. |
| LbCas12a [25] | Lachnospiraceae bacterium | 1,228 | TTTN | Similar to AsCas12a; another well-characterized Cas12a ortholog. |
| AsCas12f1 [25] | Acidaminococcus sp. | ~400-700 | NTTR | An ultra-compact nuclease, enabling new delivery options. |
Experimental Protocols for Assessing Editing Efficiency
Protocol 1: Validating On-Target Editing Using T7 Endonuclease I (T7EI) Assay
This protocol is adapted from a study optimizing CRISPR in grapevine suspension cells [28].
- Genomic DNA Extraction: Harvest transfected cells and extract genomic DNA using a standard CTAB method or commercial kit.
- PCR Amplification: Design primers flanking your target site (amplicon size 400-800 bp). Perform PCR using a high-fidelity polymerase to amplify the target region from the genomic DNA.
- DNA Denaturation and Reannealing: Purify the PCR products and quantify. Take 200-400 ng of the purified PCR product in a suitable buffer. Denature the DNA by heating to 95°C for 5-10 minutes, then slowly reanneal by ramping the temperature down to 25°C (e.g., -0.1°C/sec). This process allows the formation of heteroduplex DNA if indels are present.
- T7EI Digestion: Add T7EI enzyme to the reannealed DNA and incubate at 37°C for 15-60 minutes. The T7EI enzyme cleaves DNA at mismatches in heteroduplex molecules.
- Gel Electrophoresis: Run the digested products on a 2-3% agarose gel. Cleaved bands indicate successful editing.
- Efficiency Calculation: Use gel imaging software to quantify the band intensities.
- Editing Efficiency (%) = [1 - √(1 - (b+c)/(a+b+c))] × 100
- Where
ais the intensity of the undigested PCR product band, andbandcare the intensities of the cleaved bands.
Protocol 2: Determining Specificity Using GUIDE-seq
GUIDE-seq (Genome-wide, Unbiased Identification of DSBs Enabled by Sequencing) is an unbiased method for detecting off-target sites genome-wide [31].
- dsODN Transfection: Co-transfect your cells with the CRISPR-Cas9 plasmid(s) and a double-stranded oligodeoxynucleotide (dsODN) tag.
- Genomic DNA Extraction and Shearing: Harvest cells 2-3 days post-transfection. Extract genomic DNA and shear it to an average fragment size of 500 bp.
- Library Preparation and Enrichment:
- Blunt-end Repair & A-tailing: Prepare the sheared DNA for adapter ligation.
- Adapter Ligation: Ligate sequencing adapters to the DNA fragments.
- dsODN Enrichment: Perform PCR to enrich for fragments that have incorporated the dsODN tag.
- High-Throughput Sequencing: Sequence the resulting library on an appropriate platform (e.g., Illumina).
- Bioinformatic Analysis: Map the sequenced reads to the reference genome. Cluster reads that have the dsODN tag integrated and identify genomic locations with significant tag integration sites, which correspond to Cas9 cleavage events (both on-target and off-target).
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagent Solutions for High-Efficiency CRISPR Workflows
| Reagent / Material | Function | Example & Notes |
|---|---|---|
| High-Fidelity Cas Variants | Reduces off-target effects while maintaining on-target cleavage. | SpCas9-HF1, eSpCas9(1.1), Alt-R S.p. HiFi Cas9 [24] [25]. |
| PAM-Flexible Cas Variants | Enables targeting of genomic sites lacking canonical NGG PAMs. | xCas9, SpCas9-NG, SpRY [24]. |
| Cas Orthologs | Provides alternative PAMs and smaller sizes for delivery. | SaCas9, CjCas9, AsCas12a [25]. |
| AI-Designed Editors | Novel proteins with potentially optimized properties not found in nature. | OpenCRISPR-1 (publicly available sequence) [26]. |
| Ribonucleoprotein (RNP) Complexes | For transient, high-efficiency delivery with reduced off-target activity. | Complex of purified Cas protein and synthetic gRNA. |
| Unbiased Off-Target Detection Kits | For genome-wide identification of CRISPR-induced DSBs. | GUIDE-seq [31] & Digenome-seq [31] reagent kits. |
| HDR Donor Templates | For precise gene insertion or correction when using HDR repair. | Single-stranded oligodeoxynucleotides (ssODNs) or double-stranded DNA donors. |
Workflow Visualization: Selecting a High-Efficiency Cas Nuclease
The diagram below outlines a logical workflow for selecting the most appropriate Cas nuclease for your experiment, based on the primary experimental requirement.
Precision in Practice: Advanced Delivery Systems and Editing Platforms for Therapeutic Development
The therapeutic application of CRISPR gene editing hinges on the safe and efficient delivery of its molecular components—most commonly the Cas nuclease and a guide RNA (gRNA)—into the nucleus of target cells [32]. While the ex vivo delivery of CRISPR into cells in a culture dish is relatively straightforward, the in vivo delivery required to treat most genetic diseases presents a major challenge [33]. The delivery vehicle must protect the CRISPR machinery from degradation, navigate to the correct tissue, cross cell membranes, and release its payload effectively [32]. No single delivery modality is ideal for every application; each offers a distinct set of advantages and trade-offs concerning immunogenicity, editing duration, cargo capacity, and manufacturing scalability [32] [34].
The following diagram illustrates the core decision-making workflow for selecting and optimizing a CRISPR delivery system for therapeutic in vivo use.
Diagram 1: Decision workflow for in vivo CRISPR delivery.
Comparative Analysis of Delivery Systems
The table below provides a high-level comparison of the three primary delivery modalities to guide initial selection.
Table 1: Key Characteristics of CRISPR Delivery Systems
| Feature | Lipid Nanoparticles (LNPs) | Viral Vectors (e.g., AAV) | Virus-like Particles (VLPs) |
|---|---|---|---|
| Typical Cargo | mRNA, RNP [34] | DNA [32] | Proteins, RNPs [35] [36] |
| Immunogenicity | Low; suitable for repeated dosing [34] | High; pre-existing immunity can limit efficacy [32] [34] | Moderate; lacks viral genetic material, improving safety [36] |
| Editing Duration | Transient (days) [34] | Long-term or permanent [34] | Transient (days) [35] |
| Cargo Capacity | Moderate to High [34] | Low (packaging limit ~4.7 kb) [32] | Moderate (can deliver large proteins like Cas9) [36] |
| Manufacturing & Scalability | Highly scalable [34] | Complex and costly [34] | Complexity depends on the platform [36] |
| Primary Applications | mRNA vaccines, short-term therapies, systemic delivery [34] | Long-term gene expression for genetic disorders [34] | Transient delivery of gene-editing RNPs; combines advantages of viral and non-viral systems [35] [32] |
Troubleshooting Common Delivery Problems
FAQ: Low Editing Efficiency
Q: Despite successful cellular transduction, my CRISPR editing efficiency remains low. What are the potential causes and solutions?
- A1: Inefficient Nuclear Import: The Cas protein may not be efficiently entering the nucleus.
- Solution: Optimize nuclear localization signals (NLS). Recent research demonstrates that using hairpin internal NLS (hiNLS) sequences inserted directly into the Cas9 backbone, rather than relying solely on terminally fused NLS, can significantly enhance nuclear import and editing efficiency in primary human cells, such as T lymphocytes [3].
- A2: Cargo Mismatch or Instability:
- Solution: Match the cargo to the vector's strengths. For LNP delivery, mRNA is often more effective than plasmid DNA. For VLP delivery, ensure the ribonucleoprotein (RNP) complex is properly packaged and released. Directed evolution of VLP capsids has been used to create variants (e.g., v5 eVLP) with improved RNP packaging and post-delivery release, resulting in 2-4x higher delivery efficiency [35].
- A3: Off-target Vector Tropism: The delivery vector may not be efficiently reaching or entering your target cell type.
- Solution: Re-engineer vector targeting. For viral vectors and VLPs, this can involve engineering the envelope proteins or capsids for specific tissue tropism [36] [32]. For LNPs, the lipid composition and surface functionalization (e.g., with antibodies or peptides) can be modified to enhance target cell specificity [34].
FAQ: Cytotoxicity and Immune Response
Q: My delivery vector is causing significant cell death or triggering an immune response. How can I mitigate this?
- A1: Vector-Induced Toxicity:
- For LNPs: The lipid composition can be cytotoxic. Optimize the ratios of ionizable lipids, helper phospholipids, cholesterol, and PEG-lipids to create a more biocompatible formulation [34].
- For Viral Vectors: High viral titers can induce toxicity. Perform a dose-response curve to find the minimum effective titer. Consider switching serotypes (for AAV) or using less immunogenic viral backbones (e.g., using lentivirus instead of adenovirus where appropriate) [32] [34].
- A2: Immune Recognition:
- Solution: Use transient expression systems. The prolonged expression of Cas9 from viral vectors can trigger adaptive immune responses. Delivering pre-assembled Cas9-gRNA RNP complexes via VLPs or LNPs offers transient activity (1-2 day half-life), which minimizes both immune recognition and off-target editing risks [35] [3].
FAQ: Cargo Packaging and Release
Q: I am using VLPs, but the functional cargo is not being efficiently packaged or released in the target cell.
- A1: Inefficient Packaging:
- Solution: Fuse your cargo (e.g., a base editor protein) to the VLP scaffold protein (e.g., Gag) via a cleavable linker. Systematic optimization of this linker and the stoichiometry of packaging plasmids can dramatically enhance cargo loading [36]. Using a barcoded directed evolution approach, researchers have identified VLP capsid mutations that significantly increase RNP packaging capacity [35].
- A2: Inefficient Release in Cytosol:
- Solution: The cleavable linker between the cargo and scaffold protein is critical. Incorporate linkers that are efficiently cleaved by ubiquitous host proteases (e.g., viral proteases included in the VLP system) upon entry into the target cell's cytoplasm. Optimizing this linker sequence is a key strategy for ensuring effective cargo release and function [36].
Advanced Optimization: Experimental Protocols
Protocol: Optimizing VLP Delivery of CRISPR RNP
This protocol outlines the steps for developing and testing engineered VLPs for RNP delivery, based on recent directed evolution approaches [35].
Objective: To produce and characterize evolved VLPs (e.g., v5 eVLP) with improved production and transduction efficiencies for CRISPR-RNP delivery.
Materials:
- Producer Cells: HEK-293T cells
- Packaging Plasmids: Plasmids encoding VLP structural proteins (e.g., Gag), viral enzymes (e.g., Pol), and envelope protein (e.g., VSV-G)
- Cargo Plasmid: Plasmid encoding the cargo protein (e.g., Cas9) fused to the VLP scaffold via a cleavable linker
- Barcoded gRNA Library: A library of gRNAs where each gRNA contains a unique barcode sequence corresponding to a specific VLP variant
Method:
- Library Creation: Create a library of VLP capsid variants by introducing mutations into the gene encoding the major capsid protein.
- VLP Production: Co-transfect HEK-293T producer cells with the packaging plasmid mix and the barcoded gRNA library.
- Each VLP variant packages a gRNA with a unique barcode, linking the VLP's identity to its physical cargo [35].
- Selection Pressure: Harvest the VLPs and use them to transduce your target cell line. Apply a selective pressure (e.g., antibiotic selection if the gRNA targets an essential gene) to enrich for VLP variants that successfully deliver functional RNP.
- Variant Identification: Recover the genomic DNA from surviving target cells and sequence the barcode region of the integrated gRNA.
- The enriched barcodes identify the VLP capsid variants with the most favorable properties (e.g., high production, efficient transduction, and functional delivery) [35].
- Validation: Clone the identified beneficial mutations to create a new, optimized VLP (e.g., v5 eVLP). Validate its improved performance by comparing RNP packaging efficiency (e.g., via western blot) and functional gene editing efficiency (e.g., via T7E1 assay or NGS) against previous-generation VLPs.
Protocol: Enhancing Nuclear Delivery with hiNLS-Cas9
This protocol describes the implementation of hairpin internal nuclear localization signals (hiNLS) to boost CRISPR editing efficiency [3].
Objective: To design and test hiNLS-Cas9 constructs for enhanced editing in primary human cells.
Materials:
- hiNLS-Cas9 Constructs: Expression vectors for Cas9 with hiNLS sequences installed at selected sites within the protein backbone
- Control Constructs: Cas9 with standard terminally fused NLS
- Target Cells: Primary human T cells or other clinically relevant primary cells
- Delivery Method: Electroporation system or peptide-based non-viral delivery method
Method:
- Construct Design: Design and synthesize Cas9 genes incorporating short hiNLS sequences at internal, structured sites within the protein, avoiding catalytic domains.
- Protein Production: Express and purify the hiNLS-Cas9 and control NLS-Cas9 proteins from a suitable expression system (e.g., E. coli). Assess protein purity and yield.
- RNP Formation: Pre-complex the purified Cas9 proteins with target-specific sgRNA to form ribonucleoproteins (RNPs).
- Cell Delivery: Deliver the RNPs into primary human T cells using a clinically relevant method such as electroporation.
- Efficiency Analysis: After 2-4 days, harvest genomic DNA and assess editing efficiency at the target locus (e.g., TRAC or B2M) using T7 Endonuclease I (T7E1) mismatch detection assays or next-generation sequencing (NGS).
- Expected Outcome: hiNLS constructs should demonstrate higher editing efficiency and higher purity/yield upon production compared to terminally fused NLS constructs [3].
The Scientist's Toolkit: Essential Reagents
Table 2: Key Research Reagents for Delivery Optimization
| Reagent | Function | Example Applications |
|---|---|---|
| Ionizable Cationic Lipids | Core component of LNPs; encapsulates nucleic acids and facilitates endosomal escape [34]. | Formulating LNPs for in vivo delivery of CRISPR mRNA or sgRNA. |
| VSV-G Envelope Protein | A broad-tropism viral glycoprotein used to pseudotype viral vectors and VLPs [36]. | Enhancing the transduction efficiency of lentiviral vectors and VLPs across a wide range of cell types. |
| Cleavable Linkers (e.g., protease sites) | A peptide sequence fused between a cargo protein and a VLP scaffold; cleaved upon cell entry to release functional cargo [36]. | Enabling efficient packaging and intracellular release of editor proteins (e.g., Cas9, BE) from VLPs. |
| Hairpin Internal NLS (hiNLS) | Short peptide sequences inserted internally within the Cas9 protein to enhance nuclear import density [3]. | Boosting editing efficiency of CRISPR-Cas9 RNP in primary and hard-to-transfect cells. |
| Barcoded gRNA Library | A pool of sgRNAs containing unique nucleotide sequence "barcodes" to identify specific VLP variants [35]. | Performing directed evolution of VLP capsids to select for variants with improved delivery properties. |
| Adeno-associated Virus (AAV) | A popular viral vector known for low immunogenicity and long-term gene expression; has limited cargo capacity [32] [34]. | Delivering CRISPR components in vivo for targets that fit within its packaging limit; often used with smaller Cas orthologs like SaCas9. |
Selecting the right gene-editing platform is a critical first step in designing efficient and reliable experiments. While the classic CRISPR-Cas9 system creates double-stranded breaks (DSBs) to disrupt genes, newer platforms—Base Editing, Prime Editing, and Nickase systems—offer enhanced precision and control for specific applications. Your choice directly impacts the type of edit you can make, the precision required, and the potential for off-target effects. This guide provides a direct comparison, troubleshooting advice, and experimental protocols to help you select and optimize the ideal editor for your research goals, framed within the broader objective of maximizing editing efficiency.
Editor Comparison and Selection Guide
The table below summarizes the core characteristics of each editing platform to guide your initial selection [37] [38].
Table 1: Key Characteristics of Precision Genome Editors
| Editor Type | Mechanism of Action | Primary Applications | Key Advantages | Key Limitations |
|---|---|---|---|---|
| Nickase (Cas9 D10A) | Creates a single-strand break ("nick") in the DNA [38]. | - Gene knockout (via paired nicking)- Reducing off-target effects [38]. | - Higher specificity than wild-type Cas9; requires two guides for a DSB, reducing off-target cleavage [38]. | - Requires design and delivery of two gRNAs for efficient knockout [38]. |
| Base Editor (BE) | Uses a deaminase enzyme to directly convert one base pair to another without creating a DSB [39] [37]. | - Point mutation introduction (e.g., C•G to T•A, A•T to G•C) [37] [38].- Correcting point mutations associated with genetic disease [39]. | - No DSB avoids indel byproducts from NHEJ [39].- High efficiency and product purity [37]. | - Limited to specific base transitions; cannot make all possible point mutations, insertions, or deletions [39] [38].- Potential for off-target RNA editing. |
| Prime Editor (PE) | Uses a Cas9 nickase fused to a reverse transcriptase and a pegRNA to directly "search and replace" DNA sequences [39] [37]. | - All 12 possible point mutations [37].- Precise insertions and deletions [39] [37]. | - Versatility: Can install a wide range of edits without a DSB or donor DNA template [39].- High precision and lower off-target effects than Cas9 [39]. | - Complex pegRNA design.- Generally lower editing efficiency than base editors, requiring careful optimization [39] [37]. |
The following diagram illustrates the core mechanistic workflow common to all three editors, from component delivery to the final edited DNA sequence.
Frequently Asked Questions (FAQs)
1. When should I choose a Base Editor over a Prime Editor? Choose a Base Editor when your goal is to introduce one of the specific single-base changes it is engineered to make (e.g., C-to-T or A-to-G) and you prioritize high editing efficiency. Choose a Prime Editor when you need a type of edit that Base Editors cannot make, such as transversions (C-to-G, C-to-A, etc.), precise insertions, deletions, or any of the 12 possible point mutations [39] [37] [38].
2. How can I improve the low editing efficiency of my Prime Editor? Low prime editing efficiency is a common challenge. Focus on optimizing the prime editing guide RNA (pegRNA). This includes using a longer primer binding site (PBS) of around 13 nucleotides and ensuring the reverse transcriptase template (RTT) is not too long. Co-expressing a dominant-negative mismatch repair protein (e.g., MLH1dn) can also significantly enhance efficiency by preventing the cell from rejecting the newly incorporated edits [37].
3. My Base Editor is showing unwanted bystander edits. How can I mitigate this? Bystander edits occur when non-target bases within the activity window of the deaminase are modified. To minimize this, you can re-design your gRNA to position the target base in a location where adjacent editable bases are minimized. Alternatively, consider using engineered base editor variants with a narrower activity windows or altered sequence context preferences that are now available [37].
4. Why would I use a Nickase instead of the standard Cas9 nuclease? The primary reason is to reduce off-target effects. Because a single nick is typically repaired faithfully by the cell, using a Nickase with a single gRNA results in very low unintended mutagenesis. To create a knockout, you use a pair of nickases targeting opposite strands to generate a DSB, which requires both gRNAs to bind in close proximity, dramatically increasing specificity compared to wild-type Cas9 [38].
Troubleshooting Guides
Problem: Consistently Low Editing Efficiency Across All Editors
Potential Causes and Solutions:
- Cause 1: Inefficient nuclear delivery of editing components.
- Solution: Optimize the nuclear localization signal (NLS). A recent strategy demonstrated that using hairpin internal NLS (hiNLS) sequences within the Cas9 backbone, rather than terminally fused NLS, can enhance nuclear import and significantly boost editing efficiency in primary human cells like T lymphocytes [3].
- Cause 2: Poor gRNA or pegRNA design.
- Solution: Utilize online design tools that predict on-target activity. For pegRNAs, systematically test different PBS and RTT lengths. Always check for potential secondary structures in the guide RNA that could inhibit binding [38].
- Cause 3: Low expression or stability of the editor protein.
Problem: High Off-Target Activity with Nickase or Base Editor
Potential Causes and Solutions:
- Cause 1: Overexpression or prolonged expression of editing components.
- Cause 2: gRNA sequence has high similarity to other genomic sites.
- Cause 3 (Base Editors): gRNA-independent off-target DNA or RNA editing.
- Solution: This is due to the intrinsic activity of the deaminase domain. Use the newest generation of base editors that have been engineered with higher fidelity deaminase variants to minimize these effects [37].
Essential Experimental Protocols
Protocol 1: Designing and Testing a Prime Editing Experiment
This protocol outlines the key steps for a standard prime editing workflow in cultured cells.
1. pegRNA Design:
- Identify the target sequence and the specific edit you want to introduce.
- Design the pegRNA to include:
- spacer sequence (~20 nt) complementary to the target DNA.
- Primer Binding Site (PBS): A ~13-nucleotide sequence that binds the nicked DNA strand to initiate reverse transcription.
- Reverse Transcriptase Template (RTT): A template that encodes your desired edit(s). It should be long enough to contain the edit but minimizing length to maintain efficiency.
- Use online software to help design and score pegRNAs.
2. Component Delivery:
- Transfert your cells with a plasmid expressing the prime editor (nCas9-RT fusion) and the pegRNA, or deliver as in vitro transcribed mRNA and synthetic pegRNA. For hard-to-transfect cells, consider using viral vectors (lentivirus, AAV). The choice between all-in-one and separate vectors can impact efficiency; all-in-one vectors ensure co-expression [38].
3. Analysis of Editing Outcomes:
- Harvest genomic DNA 48-72 hours post-transfection.
- Amplify the target locus by PCR and subject the product to Sanger sequencing. For a more quantitative measure, use high-throughput next-generation sequencing (NGS) to calculate the precise percentage of alleles edited [40].
Protocol 2: Evaluating Editor Specificity with Tag-Seq
This scalable method accurately identifies and characterizes editor-induced double-strand breaks or unintended edits across the genome [40].
1. Editor Transfection: Introduce your editing construct (e.g., Base Editor, Prime Editor) into your cell line (e.g., HEK293T, MCF7) using your standard method.
2. Genomic DNA Extraction and Shearing: Harvest cells and extract high-quality genomic DNA. Fragment the DNA by sonication or enzymatic digestion to an average size of 300-500 bp.
3. Library Preparation and Sequencing:
- Ligate sequencing adapters to the fragmented DNA.
- Enrich for the target regions of interest or perform whole-genome sequencing.
- Sequence the libraries on an appropriate NGS platform.
4. Data Analysis:
- Map the sequencing reads to the reference genome.
- Use specialized analysis pipelines (e.g., the Tag-seq pipeline) to identify significant clusters of reads with indels (for nickase) or single-nucleotide variants (for base editors) that indicate potential off-target sites [40].
- Compare the experimental group to an untreated control to filter out background noise.
The Scientist's Toolkit: Essential Research Reagents
The table below lists key materials required for setting up and analyzing genome editing experiments.
Table 2: Essential Reagents for Genome Editing Experiments
| Reagent / Material | Function / Application | Example & Notes |
|---|---|---|
| Editor Expression Plasmid | Delivers the genetic code for the editor protein (e.g., BE, PE, Nickase) into the cell. | Plasmids like pCMV-PE2 for prime editing; all-in-one vectors simplify delivery [38]. |
| Guide RNA Expression Vector | Delivers the code for the gRNA or pegRNA. | Can be on a separate plasmid or part of an all-in-one system. U6 promoter is commonly used [38]. |
| Delivery Reagents | Facilitates the entry of editing components into cells. | Lipofectamine 3000 for plasmid transfection; electroporation for RNP delivery [38]. |
| Ribonucleoprotein (RNP) Complex | Pre-complexed Cas protein and gRNA for transient, highly specific editing. | Ideal for sensitive primary cells (e.g., T-cells); reduces off-target effects and immune stimulation [3] [38]. |
| Donor DNA Template | Provides the homology template for HDR (for Nickase-mediated knock-in) or as a reference for design. | Single-stranded oligodeoxynucleotide (ssODN) for small edits; double-stranded DNA for larger insertions [38]. |
| Genomic DNA Isolation Kit | Purifies DNA from cells for genotyping analysis. | Essential for post-editing validation (e.g., T7EI assay, Sanger sequencing, NGS). |
| PCR Reagents | Amplifies the target genomic locus for analysis. | High-fidelity polymerases are recommended to avoid introducing errors during amplification. |
| Next-Generation Sequencing Kit | For deep, quantitative analysis of on-target and off-target editing. | Provides the most comprehensive data on editing efficiency and specificity (e.g., Tag-seq) [40]. |
FAQs and Troubleshooting Guides
Neurons
Q: How can I improve neuronal survival during CRISPR screening? A: Neurons are particularly sensitive to DNA damage. To enhance survival and editing efficiency, consider switching from CRISPRn (CRISPR knockout) to CRISPRi (CRISPR interference). CRISPRi uses a catalytically dead Cas9 (dCas9) to silence gene expression without causing DNA double-strand breaks, which is less toxic to post-mitotic cells like neurons [41]. Furthermore, when performing survival-based screens, ensure the use of a focused, relevant library (e.g., targeting kinases or drug targets) to reduce stress on the cells from excessive transduction [41].
Q: What screening readouts work best for neuronal phenotypes? A: Neuronal function involves complex phenotypes like morphology and transcriptomic state. You can effectively screen for these using:
- High-content imaging to quantify changes in neurite outgrowth and branching [41].
- Single-cell RNA sequencing (e.g., CROP-seq, Perturb-seq) to profile transcriptomic changes resulting from genetic perturbations [41].
- FACS-based sorting if the phenotype can be linked to a fluorescent reporter or antibody staining [41].
Cardiomyocytes
Q: What is an efficient method to deliver CRISPR reagents to human iPSC-derived cardiomyocytes? A: A major challenge is achieving high editing efficiency without compromising cell viability or differentiation potential. An optimized protocol is to perform reverse transduction at the iPSC stage [42].
- Protocol: Mix the lentiviral CRISPR library with dissociated iPSCs during plating, rather than adding the virus to already-adherent cells. This dramatically increases infection efficiency by exposing more cell surface area to the viral particles [42].
- Key Control: After transduction and selection, always check that your iPSCs maintain pluripotency markers (OCT4, Nanog, SOX2) and that they can still successfully differentiate into rhythmically beating cardiomyocytes expressing markers like cardiac Troponin T (cTNT) [42].
Q: How can I model specific disease toxicities, like chemotherapy-induced cardiotoxicity, in cardiomyocytes? A: Genome-wide CRISPR knockout screens in iPSC-derived cardiomyocytes are powerful for this. As a proof of concept:
- Differentiate library-transduced iPSCs into cardiomyocytes [42].
- Treat the cardiomyocytes with the drug of interest (e.g., Doxorubicin) to induce cell death [42].
- Sequence the sgRNAs in surviving cardiomyocytes to identify "hits" – genes whose knockout conferred resistance. This approach has successfully identified novel human-specific transporters like SLCO1A2 that protect against Doxorubicin cardiotoxicity [42].
Primary Cells & Hard-to-Transfect Cells
Q: How can I enhance CRISPR editing efficiency in primary human T cells for therapeutic applications? A: For primary immune cells like T cells, ribonucleoprotein (RNP) delivery is highly effective. To further boost efficiency, recent research has engineered hairpin internal Nuclear Localization Signals (hiNLS) within the Cas9 protein [3].
- Mechanism: These hiNLS constructs are integrated into the Cas9 backbone, unlike traditional terminally-fused NLS tags. This increases the density of NLS signals, improving nuclear import of the Cas9 RNP complex [3].
- Benefit: Since RNP has a short half-life, rapid nuclear entry is critical for high editing rates before the complex is degraded. This strategy has shown enhanced knockout of genes like B2M and TRAC in primary human T cells, which is crucial for cell-based therapies [3].
Q: My screen in a complex primary-like model (e.g., an organoid) has poor representation. How can I maintain library complexity? A: Maintaining library complexity through differentiation is critical. Start with a large number of iPSCs (e.g., 90 million) for library transduction to ensure initial diversity [42]. After puromycin selection and a period of expansion, sequence the sgRNAs to confirm minimal loss (e.g., <10% of sgRNAs). Successful differentiation should retain near genome-wide representation, with at least 3 sgRNAs per gene targeting over 13,000 genes in the final differentiated cell type [41] [42].
Table 1: Overview of CRISPR Screening Strategies in Different Cell Types
| Cell Type | CRISPR Type | Example Screening Phenotype | Library Size / Type | Key Challenge Addressed |
|---|---|---|---|---|
| Neurons [41] | CRISPRi | Neuronal survival, neurite morphology | ~2,000 genes (focused) | Sensitivity to DNA damage |
| Cardiomyocytes [42] | CRISPRn | Doxorubicin-induced cardiotoxicity | ~75,000 sgRNAs (genome-wide) | Low viral infection efficiency |
| Neural Progenitors [41] | CRISPRn | Susceptibility to Zika virus infection | Genome-wide | Viral infection & cell survival |
| Microglia [41] | CRISPRi/a | Phagocytosis, cell activation | ~2,000 genes (focused) | Complex functional phenotypes |
| T Cells (Primary) [3] | CRISPRn (RNP) | Knockout efficiency for therapy | N/A (specific targets) | Low editing efficiency in primary cells |
Table 2: Experimental Protocol Summary from Key Studies
| Experimental Step | Neurons (Tian et al.) [41] | Cardiomyocytes (Sapp et al.) [42] | Primary T Cells (Wilson et al.) [3] |
|---|---|---|---|
| CRISPR Format | CRISPRi (dCas9 repressor) | CRISPRn (Lentiviral library) | CRISPRn (hiNLS-Cas9 RNP) |
| Delivery Method | Lentiviral transduction | Reverse lentiviral transduction | Electroporation |
| Key Reagents | CRISPRi-v2 library | Whole-genome CRISPRko library | hiNLS-Cas9 RNP complex |
| Screening Readout | Single-cell RNA-seq, Imaging | Cell survival under drug selection | Flow cytometry for protein knockout |
| Key Finding | Genes regulating survival & morphology | SLCO1A2 loss protects from toxicity | Enhanced editing efficiency & yield |
Experimental Workflows
The Scientist's Toolkit: Key Research Reagent Solutions
Table 3: Essential Reagents and Tools for Cell-Type Specific CRISPR Screening
| Reagent / Tool | Function | Application Context |
|---|---|---|
| CRISPRi System [41] | dCas9 fused to transcriptional repressor (KRAB); silences genes without DNA cleavage. | Ideal for neurons and other DNA damage-sensitive cells. |
| hiNLS-Cas9 [3] | Cas9 variant with hairpin internal Nuclear Localization Signals; enhances nuclear import. | Improves editing efficiency in primary cells (T cells) with RNP delivery. |
| Whole-genome CRISPRko Library [42] | Pooled lentiviral library (e.g., ~75,000 sgRNAs) for genome-wide knockout screens. | For unbiased discovery screens in iPSC-derived cells (cardiomyocytes). |
| Focused Library [41] | Smaller library targeting specific gene families (kinases, drug targets). | Reduces stress on cells; ideal for complex models like neurons and organoids. |
| CRISPR-GRANT [43] | Stand-alone graphical tool for CRISPR indel analysis from NGS data. | User-friendly analysis for wet-lab researchers; no command-line needed. |
| CRISPRMatch [44] | Automated pipeline for calculating mutation frequency and editing efficiency. | Processing high-throughput genome-editing data from NGS. |
High-Throughput Screening (HTS) is a powerful method used in drug discovery and the development of therapeutic agents that involves automated equipment to rapidly test thousands of samples [45]. In the context of CRISPR gene editing, HTS enables large-scale functional characterization of genes and rapid identification of optimal genetic targets [46]. This approach represents a significant advancement over single-gene editing experiments, allowing researchers to investigate complex biological processes and genetic interactions systematically [47]. The flexibility and editing efficiency of CRISPR/Cas9 systems have established them as leading tools for high-throughput genetic screening, overcoming limitations of previous technologies like RNA interference (RNAi) which often resulted in incomplete gene suppression and high off-target effects [46].
This technical support center provides comprehensive guidance on implementing high-throughput screening systems to characterize gene editors rapidly, with particular emphasis on optimizing editing efficiency—a crucial consideration for both basic research and clinical translation [3].
Core Concepts and Methodologies
Understanding Screening Approaches
High-throughput genetic screening primarily encompasses two complementary approaches:
- Loss-of-Function (LOF) Screening: Aims to silence or knock out genes to identify sensitive points in biological pathways. Traditional CRISPR/Cas9 knockout screening falls into this category [46].
- Gain-of-Function (GOF) Screening: Utilizes CRISPR-mediated gene activation (CRISPRa) methods for temporary gene transcription activation to study gene overexpression effects [46].
Essential High-Throughput Workflow
The following diagram illustrates the generalized workflow for conducting high-throughput CRISPR screens:
Figure 1: High-throughput screening workflow for CRISPR editor characterization.
Experimental Protocols
Next-Generation Sequencing Screening Protocol
This protocol enables parallel screening of up to 96 clones using next-generation sequencing (NGS) to detect CRISPR-Cas9 induced mutations [48].
Day 1: Preparation
- Design and synthesize sgRNAs targeting your gene of interest using validated design tools [49].
- Select appropriate CRISPR format: purified transfection-ready gRNA libraries or lentiviral CRISPR particles [47].
- Prepare recipient cells (e.g., mouse embryonic stem cells, primary T cells, or other clinically relevant cell types).
Day 2: Transfection/Transduction
- Deliver CRISPR components using optimized methods:
- Electroporation: For ribonucleoprotein (RNP) complexes, especially in primary cells [3].
- Lentiviral transduction: For stable integration, using controlled titer (>10⁶ TU/ml) [47].
- Peptide-enabled RNP delivery: A hardware-free, nonviral approach serving as a proxy for in vivo delivery technologies [3].
- Include appropriate controls (non-targeting sgRNAs, mock transfected).
Day 3-7: Selection and Expansion
- Apply selection pressure if using antibiotic resistance markers.
- Expand transfected cell pools for 3-5 days to allow editing and phenotype manifestation.
Day 8: Screening and Analysis
- Harvest cells and extract genomic DNA.
- Amplify target regions by PCR. For problematic GC-rich regions, add 1-10 μL of GC Enhancer in a 50 μL reaction [30].
- Perform next-generation sequencing (Ion Torrent PGM or equivalent platform).
- Analyze sequencing data to identify mutated clones, including homozygous, heterozygous, and mixed populations [48].
Advanced Nuclear Localization Enhancement Protocol
To optimize editing efficiency in therapeutically relevant primary cells (e.g., human lymphocytes):
- Engineer Cas9 constructs with hairpin internal Nuclear Localization Signals (hiNLS) at selected sites within the backbone [3].
- Compare hiNLS constructs against terminally fused NLS sequences as controls.
- Deliver as RNP complexes via electroporation to maintain transient exposure [3].
- Evaluate editing efficiency at target genes (e.g., B2M, TRAC) 48-72 hours post-delivery.
- Assess production yield and purity to ensure manufacturing scalability [3].
Troubleshooting Guides
Common Experimental Challenges and Solutions
| Problem | Possible Cause | Solution |
|---|---|---|
| Low editing efficiency [3] [30] | Suboptimal nuclear localization | Implement hiNLS constructs to enhance nuclear import [3] |
| Low transfection efficiency | Optimize delivery method; use Lipofectamine 3000 or 2000 reagent [30] | |
| Cell type-dependent limitations | Use 293FT cells as a positive control; optimize for primary cells [30] | |
| High off-target effects [30] | Poorly designed crRNA oligos | Carefully design crRNA targets to avoid homology with other genomic regions [30] |
| No cleavage band visible [30] | Nucleases cannot access target | Design new targeting strategy for nearby sequences [30] |
| Transfection efficiency too low | Optimize transfection protocol [30] | |
| Smear on gel [30] | Lysate too concentrated | Dilute lysate 2- to 4-fold and repeat PCR [30] |
| Too faint PCR product [30] | Lysate too dilute | Double amount of lysate in PCR reaction (max 4 μL) [30] |
| No PCR product [30] | Poor primer design or GC-rich region | Redesign primers (18-22 bp, 45-60% GC content, Tm 52-58°C) [30] |
| Disparity in band intensity [30] | Varying lysate concentrations | Purify PCR products and use same DNA quantity (50-100 ng) per reaction [30] |
Advanced Troubleshooting: Specialized Applications
Primary Cell Screening Challenges
- Problem: Limited editing efficiency in primary human T cells
- Solution: Utilize hiNLS Cas9 variants in RNP format delivered via electroporation [3]
- Validation: Assess knockout efficiencies for B2M and TRAC genes as benchmarks [3]
In Vivo Delivery Challenges
- Problem: inefficient delivery for therapeutic applications
- Solution: Implement lipid nanoparticle (LNP) delivery systems that accumulate in target organs (particularly liver) [12]
- Advanced Approach: Consider redosing strategies possible with LNP (not feasible with viral vectors) [12]
Frequently Asked Questions (FAQs)
What are the main advantages of CRISPR screening over RNAi-based approaches? CRISPR screening provides more complete and permanent gene disruption, resulting in clearer phenotypes and fewer false readouts compared to the variable knock-down achieved with RNAi. CRISPR libraries also offer greater design flexibility and can target non-coding genomic regions [46] [47].
How can I improve editing efficiency in difficult-to-transfect primary cells? Focus on nuclear localization enhancement strategies. Recent research demonstrates that incorporating hairpin internal Nuclear Localization Signals (hiNLS) within the Cas9 backbone significantly improves editing efficiency in primary human lymphocytes compared to traditional terminally fused NLS sequences [3].
What factors should I consider when choosing between arrayed and pooled screening formats? Arrayed libraries (e.g., in 96-well format) allow individual well analysis without deconvolution, while pooled screens require sequencing for target identification. Arrayed formats work well with automated systems and are ideal for comprehensive phenotypic analysis, whereas pooled screens enable larger library sizes but require more complex downstream analysis [47].
How do I address low efficiency in my CRISPR screening workflow? Consider these strategies: (1) Add antibiotic selection or FACS sorting to enrich transfected cells; (2) Optimize delivery method - use high-quality RNP complexes or lentiviral particles with appropriate titer; (3) Implement NLS engineering to enhance nuclear import [3] [30].
What methods are available for detecting CRISPR-induced mutations in high-throughput formats? Next-generation sequencing platforms (e.g., Ion Torrent PGM) enable parallel screening of up to 96 clones simultaneously. This approach identifies homozygous, heterozygous, and mixed clones while providing information on clonal heterogeneity [48].
Can I perform high-throughput screening in non-dividing or primary cells? Yes, lentiviral vectors can transduce both dividing and non-dividing cells. For primary cells like T cells, optimized RNP delivery via electroporation has proven effective, especially with enhanced NLS constructs [3] [47].
Research Reagent Solutions
The following table summarizes essential materials for establishing high-throughput CRISPR screening workflows:
| Reagent/System | Function | Application Notes |
|---|---|---|
| Arrayed Purified gRNA Libraries [47] | Pre-synthesized, transfection-ready guides | >97% yield for most targets; normalized concentrations; 96-well format |
| Lentiviral CRISPR Particles [47] | Stable delivery of CRISPR components | Titer >10⁶ TU/ml; suitable for dividing & non-dividing cells |
| Cas9 Nuclease Protein [47] | Ready-to-use enzyme for RNP complexes | High-purity, transfection-grade; compatible with gRNA libraries |
| hiNLS Cas9 Variants [3] | Enhanced nuclear import | Improved editing efficiency in primary cells; maintained high yield production |
| GeneArt Genomic Cleavage Detection Kit [30] | Detect CRISPR-mediated cleavage | Verification of cleavage on endogenous genomic loci |
| 384-well Nucleofector System [45] | High-throughput electroporation | Integrates with LHS systems (Tecan, Beckman, Hamilton); ideal for primary cell screens |
Advanced Applications and Future Directions
The following diagram illustrates the strategic integration of high-throughput screening in therapeutic development:
Figure 2: High-throughput screening in therapeutic development workflow.
High-throughput CRISPR screening has become a cornerstone technology in functional genomics and therapeutic development [45]. As the field advances, several promising directions are emerging:
- Multiplexed Editing: Libraries utilizing combinations of four gRNAs (4gRNA-combo) enable simultaneous targeting of multiple gene pairs within single cells [46].
- Advanced Editor Platforms: Base editors (BEs) and epigenetic editors that don't rely on double-strand breaks offer solutions to technological challenges in screening [46].
- Therapeutic Translation: High-throughput screening identifies viable gene targets for enhancement or inhibition, potentially addressing challenges in cellular therapies like CAR-T cell exhaustion and antigen escape [46].
The continued refinement of high-throughput screening methodologies will play a crucial role in optimizing CRISPR editing efficiency and accelerating the development of next-generation genomic medicines.
Solving Efficiency Challenges: Practical Strategies for Enhanced On-Target Editing
Troubleshooting Common sgRNA Design and Experimentation Problems
This guide addresses frequent challenges encountered during CRISPR gene editing experiments, providing targeted solutions to help researchers achieve optimal efficiency and specificity.
Q1: My CRISPR experiment has low editing efficiency. What could be wrong and how can I fix it?
Low editing efficiency is often related to sgRNA design or delivery. To resolve this, first verify your sgRNA sequence. Ensure it is 17-23 nucleotides long and has a GC content between 40% and 60% [50]. Consecutive nucleotide repeats, such as poly-T or poly-G tracts, can hinder efficiency and should be avoided [50] [51]. Second, confirm that your delivery method (e.g., plasmid, RNP, viral vector) is effective for your specific cell type, as optimization may be required [9]. Finally, check the chromatin accessibility of your target region; sites in open chromatin are generally more accessible and lead to higher efficiency. Using chromatin accessibility data (e.g., from ATAC-seq) can guide target selection [52].
Q2: How can I minimize off-target effects in my gene editing workflow?
Minimizing off-target effects is critical for experimental validity. Start by using computational tools to design highly specific sgRNAs. Tools like CHOPCHOP, E-CRISP, and Cas-OFFinder can predict potential off-target sites by scanning the entire genome for sequences with partial complementarity to your sgRNA [53]. You should prioritize sgRNA candidates with minimal sequence similarity to other genomic regions, especially in coding sequences [52] [51]. Furthermore, consider using high-fidelity Cas9 variants (e.g., SpCas9-HF1) that have been engineered to reduce off-target cleavage while maintaining on-target activity [9].
Q3: I suspect mosaicism in my edited cell population. How can I address this?
Mosaicism, where a mixture of edited and unedited cells exists, can be addressed by optimizing the timing of CRISPR component delivery. Ensure delivery is synchronized with the cell cycle stage of your target cells [9]. Using inducible Cas9 systems can provide more control over the timing of editing. To isolate a fully edited cell line, techniques such as single-cell cloning or dilution cloning are recommended after the editing process [9].
Q4: My experiment resulted in unexpected cell toxicity. What might be the cause?
Cell toxicity can occur due to high concentrations of CRISPR-Cas9 components, particularly when using plasmid DNA [9]. To mitigate this, optimize the concentration of your delivered sgRNA and Cas9. Start with lower doses and titrate upwards to find a balance between effective editing and cell viability [9]. As an alternative, consider delivering preassembled Cas9 ribonucleoprotein (RNP) complexes, which can lead to faster editing with reduced off-target effects and potentially lower cytotoxicity [50].
If you cannot detect edits, first confirm that your genotyping method is sufficiently sensitive. Robust methods include T7 Endonuclease I (T7E1) assays, Surveyor assays, or next-generation sequencing (NGS) [51] [9]. Second, ensure your transfection efficiency is high enough; low efficiency can mean too few cells received the editing components. Optimize your transfection protocol or use methods like antibiotic selection or FACS sorting to enrich for transfected cells [54]. Finally, always include proper controls—a positive control with a validated sgRNA and a negative control with a non-targeting sgRNA—to benchmark performance and account for background noise [9].
Frequently Asked Questions (FAQs) on sgRNA Design
Q: What is the ideal length and GC content for a standard SpCas9 sgRNA? A: The ideal sgRNA length is typically 20 nucleotides for the guide sequence. The GC content should be balanced, ideally between 40% and 60%, to ensure stable binding without promoting excessive rigidity or off-target effects [52] [50] [51].
Q: How does the PAM requirement influence sgRNA design? A: The Protospacer Adjacent Motif (PAM) is essential for Cas protein recognition and cleavage. For the commonly used SpCas9, the PAM sequence is 5'-NGG-3', located immediately downstream of the target site on the non-complementary DNA strand. Different Cas proteins (e.g., SaCas9, Cas12a) recognize different PAM sequences, which directly determines where in the genome you can design your sgRNA [52] [50].
Q: Why is it important to consider sgRNA secondary structure, and how can I analyze it? A: Strong secondary structures, such as hairpins, in the sgRNA can prevent it from binding effectively to the target DNA, significantly reducing editing efficiency [52]. You can use computational tools to predict secondary structures during the design phase, allowing you to select sequences that minimize such unfavorable conformations [52].
Q: What are some advanced strategies to enhance sgRNA performance? A: Beyond basic design rules, strategies include using chemically modified synthetic sgRNAs to improve stability and protect from degradation [50]. Another approach is the rational engineering of the sgRNA scaffold, such as adding a hairpin structure to prevent misfolding at difficult-to-edit sites [50].
Q: Are there AI tools that can assist with the entire experimental design process? A: Yes, tools like CRISPR-GPT have been developed to act as an AI co-pilot. They can help generate experimental designs, predict off-target edits, and troubleshoot design flaws by leveraging vast amounts of published data, making the process accessible even to those less familiar with gene editing [55].
Quantitative Data for sgRNA Design
The following table summarizes key numerical parameters to consider for optimal sgRNA design, primarily for the SpCas9 system.
| Parameter | Optimal Range or Value | Rationale & Consequences |
|---|---|---|
| sgRNA Length | 17-23 nucleotides (typically 20 nt) [50] | Longer sequences may increase off-target editing; shorter sequences compromise specificity [50]. |
| GC Content | 40-60% [52] [50] [51] | Ensures stable binding; low GC reduces stability, high GC can cause sgRNA rigidity and off-target effects [52] [50]. |
| PAM Sequence (SpCas9) | 5'-NGG-3' | Essential for Cas9 recognition and DNA cleavage; must be present immediately downstream of the target site [52] [50]. |
Experimental Protocols for sgRNA Validation
Protocol 1: Assessing sgRNA Cleavage Efficiency Using a T7 Endonuclease I (T7E1) Assay
- Extract Genomic DNA: Harvest genomic DNA from transfected or transduced cells 48-72 hours post-delivery.
- PCR Amplification: Amplify the target genomic region using specific primers that flank the edited site.
- DNA Denaturation and Renaturation: Purify the PCR product and subject it to a denaturation and reannealing process using a thermocycler. This allows the formation of heteroduplex DNA (mismatched duplexes from annealed wild-type and edited strands) in addition to homoduplexes.
- T7E1 Digestion: Digest the reannealed DNA with the T7 Endonuclease I enzyme, which cleaves at mismatched sites in heteroduplex DNA.
- Analysis: Run the digested products on an agarose gel. Cleaved DNA bands indicate the presence of successful edits, and the intensity of these bands relative to the undigested band can be used to estimate the indel frequency [51] [9].
Protocol 2: Confirming Edits and Assessing Mosaicism by Sequencing
- Clone PCR Products: After amplifying the target region (as in Protocol 1, step 2), clone the PCR products into a sequencing vector.
- Pick Individual Colonies: Pick numerous single bacterial colonies and culture them separately to obtain clonal plasmid DNA.
- Sanger Sequencing: Sequence a sufficient number of clones (e.g., 20-50) from the edited cell population.
- Sequence Alignment: Align the sequences from individual clones to the wild-type sequence. This method not only confirms the presence of edits but also quantifies the degree of mosaicism by revealing the exact variety and distribution of indel mutations within the cell population [9].
Research Reagent Solutions
The table below lists essential materials and reagents used in sgRNA-based CRISPR experiments.
| Reagent / Tool | Function / Application |
|---|---|
| U6 Promoter Plasmids | High-expression vectors for transcribing sgRNA within host cells [50]. |
| Cas9 Nuclease (WT & HiFi) | Enzyme that creates double-strand breaks in DNA; high-fidelity (HiFi) variants reduce off-target effects [9]. |
| Synthetic sgRNA | Chemically synthesized guide RNA; offers rapid action and can be modified to enhance stability [50]. |
| Ribonucleoprotein (RNP) Complexes | Preassembled complexes of Cas9 protein and sgRNA; enable fast, precise editing with minimal off-target activity [50]. |
| Lipofectamine 3000/2000 | Lipid-based transfection reagents for delivering CRISPR components into a wide range of cell types [54]. |
| T7 Endonuclease I (T7E1) Kit | Kit for detecting indel mutations and estimating editing efficiency via enzymatic mismatch cleavage [51]. |
| CHOPCHOP / E-CRISP / Benchling | Web-based bioinformatics tools for designing and predicting the efficiency and specificity of sgRNAs [53]. |
Visual Guide to the sgRNA Optimization Workflow
The following diagram illustrates the key decision points and checks in the optimal sgRNA design process.
sgRNA Design Optimization Workflow
Visualizing the Impact of Secondary Structures
This diagram conceptualizes how a stable secondary structure within the sgRNA itself can interfere with its binding to the target DNA.
Impact of sgRNA Secondary Structure
FAQs and Troubleshooting Guides
Frequently Asked Questions (FAQs)
Q1: What are the primary factors that influence transfection efficiency? Transfection efficiency is influenced by several critical factors: the type and health of the cells being transfected (low passage numbers are preferable), the chosen transfection method (chemical vs. physical), the quality and concentration of the nucleic acid used, and overall cell culture conditions, including confluency and the absence of contaminants like mycoplasma [56] [57] [58].
Q2: Why does CRISPR-Cas9 gene editing sometimes fail, and how can this be prevented? CRISPR fails about 15% of the time because the Cas9 nuclease can remain persistently bound to the DNA at the cut site after making a double-strand break. This blocks the cell's DNA repair machinery from accessing and repairing the DNA. To prevent this, design your single-guide RNA (sgRNA) so that it anneals to the DNA's template strand. This allows translocating RNA polymerases to collide with and dislodge the stalled Cas9, freeing the DNA for repair and significantly boosting editing efficiency [59] [60].
Q3: My transfection efficiency is low, but my nucleic acid quality is good. What else should I check? If your nucleic acid is high-quality, investigate your transfection complex formation. Ensure you are using serum-free medium for dilution and complexing, as serum can inhibit formation. Verify that the ratio of DNA (µg) to transfection reagent (µl) is optimal for your specific cell line—common ratios range from 1:1 to 1:5. Also, confirm that your culture medium does not contain antibiotics or other inhibitors like dextran sulfate during the transfection process [57] [58].
Q4: How can I accurately measure transfection efficiency beyond protein expression? For a direct measure of nucleic acid uptake independent of expression, you can label your plasmid DNA with a fluorophore using products like the Label IT Tracker kit before transfection. The intracellular fluorescence from the labeled DNA can then be quantified using flow cytometry. This method is particularly useful for applications like CRISPR where successful delivery does not always result in immediate protein expression [61] [62].
Q5: What are the latest advancements in delivery systems for improving cellular uptake? Recent developments include lipid nanoparticle spherical nucleic acids (LNP-SNAs). These nanostructures wrap CRISPR tools in a protective, dense shell of DNA. This architecture is recognized by cell surface receptors, promoting active cellular uptake and enhancing endosomal escape. This system has been shown to triple gene-editing efficiency and significantly reduce toxicity compared to standard lipid nanoparticles [13].
Troubleshooting Common Issues
Problem: Low Transfection Efficiency Low efficiency can halt experiments. The table below summarizes common causes and their solutions.
| Cause | Solution |
|---|---|
| Degraded or poor-quality DNA | Verify DNA integrity via A260/A280 spectrophotometry (ratio ≥1.7) and gel electrophoresis [57]. |
| Suboptimal DNA:Reagent ratio | Titrate the ratio of DNA (µg) to transfection reagent (µl), typically between 1:1 and 1:5, to find the optimum for your cell line [58]. |
| Incorrect cell density | Transfect cells when they are 70-90% confluent for most protocols. Some reagents, like Lipofectamine 2000, perform best at >90% confluency [57] [58]. |
| Serum during complex formation | Always use serum-free medium (e.g., Opti-MEM) to dilute the DNA and transfection reagent and to form complexes [57] [58]. |
| Culture contaminants | Test cells for mycoplasma and avoid using antibiotics during the transfection process itself [57]. |
Problem: Low Cell Viability After Transfection Cellular toxicity is a major hurdle. The following table outlines key culprits and how to address them.
| Cause | Solution |
|---|---|
| Endotoxin contamination in DNA | Use high-quality plasmid purification kits (e.g., endotoxin-free kits) to prepare your DNA [58]. |
| Suboptimal DNA:Reagent ratio | An excessive amount of cationic transfection reagent is toxic. Re-optimize the ratio to find a less toxic balance [57] [58]. |
| Low cell density at transfection | Ensure cells are at a healthy density (e.g., 70-90% confluent) to withstand the stress of transfection [57]. |
| Antibiotics in medium | Never include antibiotics like penicillin-streptomycin in the culture medium during transfection, as it increases permeability and toxicity [58]. |
| Improper reagent storage | Store transfection reagents at 4°C as recommended. Do not freeze or store at room temperature for extended periods, as this can degrade their function and increase toxicity [57] [58]. |
Experimental Protocols
Protocol 1: Flow Cytometric Determination of Transfection Efficiency
This protocol allows for the simultaneous quantification of nucleic acid uptake, protein expression, and cell viability [61].
Key Research Reagent Solutions
- DNA Label IT Tracker (FITC): Used to covalently label plasmid DNA with a fluorophore for tracking uptake.
- TransIT-X2 / Lipofectamine 2000 / Jet Prime: Examples of chemical transfection reagents for forming DNA complexes.
- Ghost Violet 450 Live/dead dye: A viability dye to distinguish live from dead cells in the assay.
- Antibody conjugated to CF647 dye: For detecting specific protein expression post-transfection.
Methodology
- Plasmid Labeling: The day before transfection, label your plasmid DNA (e.g., pNL4-3, pUltraHot) using the Label IT Tracker kit. Use 0.5 µl of FITC label per 1 µg of DNA. Remove unbound label via ethanol precipitation and check the final concentration and purity with a spectrophotometer [61].
- Cell Transfection: Plate your cells (e.g., 293T, Jurkat) to reach 70-80% confluency at the time of transfection.
- For adherent cells (293T): Transfect with 1 µg of labeled plasmid DNA using an appropriate reagent (e.g., 3 µl TransIT-X2 in serum-free media). Replace with complete media 6 hours post-transfection [61].
- For suspension cells (Jurkat): Electroporate 1 x 10^6 cells with 4 µg of labeled plasmid using an exponential protocol (e.g., 250 V, 350 µF) [61].
- Cell Harvest and Staining: Harvest cells 24 hours post-transfection.
- Wash cells with PBS.
- Fix cells with 4% PFA for 15 minutes at room temperature.
- Permeabilize cells with 0.2% Tween/PBS for 15 minutes.
- For protein detection, stain cells with a fluorescently conjugated antibody against your target protein (e.g., anti-p24-CF647) for 30 minutes at 4°C [61].
- Flow Cytometry Analysis: Analyze the fixed cells on a flow cytometer. Use the following gating strategy to determine efficiency:
- Uptake Efficiency: The percentage of cells positive for the FITC signal (from the labeled DNA).
- Protein Expression Efficiency: The percentage of cells positive for the antibody signal (e.g., CF647).
- Viability: The percentage of cells excluding the live/dead dye [61].
Protocol 2: A Genome-Scale CRISPR Screen for Modifiers of Cellular LDL Uptake
This protocol outlines a screening strategy to identify genetic modifiers of a specific cellular function using CRISPR [63].
Methodology
- Library Transduction: Transduce approximately 25 million HuH7 cells (a hepatocyte model) with a pooled lentiviral GeCKOv2 library, which delivers Cas9 and about 123,411 gRNAs targeting nearly all human protein-coding genes [63].
- Cell Expansion and Selection: Allow the transduced cells to expand in culture for 13 days to enable gene editing and turnover of wild-type proteins. Split the cells and culture them for one additional day under either lipoprotein-rich or lipoprotein-depleted conditions to modulate LDLR expression [63].
- Phenotypic Sorting: Incubate the mutant cells with fluorescently-conjugated LDL for 1 hour. Use a flow cytometer to sort the population into two bins: the top 7.5% (LDL${high}$) and the bottom 7.5% (LDL${low}$) of LDL-uptaking cells [63].
- Genomic DNA Extraction and Sequencing: Isolate genomic DNA from each sorted population. Amplify the integrated gRNA sequences by PCR and subject them to next-generation sequencing to determine the relative abundance of each gRNA in the LDL${high}$ versus LDL${low}$ populations [63].
- Data Analysis: Use bioinformatic tools (e.g., MAGeCK) to identify gRNAs and genes that are statistically enriched or depleted in either bin. These represent negative and positive regulators of LDL uptake, respectively [63].
Data Presentation
Quantitative Data on Transfection and CRISPR Efficiency
The following table consolidates key quantitative findings from recent research on improving delivery and editing efficiency.
| Metric | Standard Method Performance | Improved Method Performance | Method / Cause of Improvement |
|---|---|---|---|
| CRISPR Failure Rate | ~15% [59] [60] | Can be significantly reduced [60] | Designing sgRNA to anneal to the template strand for RNA polymerase-mediated Cas9 dislodging [59] [60]. |
| Cellular Uptake | Baseline (standard LNPs) [13] | Increased by up to 3x [13] | Using Lipid Nanoparticle Spherical Nucleic Acids (LNP-SNAs) as a delivery vehicle [13]. |
| Gene-Editing Efficiency | Baseline (standard delivery) [13] | Tripled (3x) [13] | Delivery via LNP-SNAs [13]. |
| Precise DNA Repair Success | Baseline [13] | Increased by >60% [13] | Delivery via LNP-SNAs [13]. |
| Critical DNA Quality Metric | A260/A280 ratio ≥ 1.7 [57] | - | Confirm DNA purity via spectrophotometry [57]. |
Visualizations
Diagram: Mechanism of CRISPR-Cas9 Failure and Rescue
The diagram below illustrates how persistent Cas9 binding blocks DNA repair and how RNA polymerase collision can rescue editing efficiency.
Diagram: Workflow for Genome-Scale CRISPR Screening
This workflow outlines the key steps in a functional CRISPR screen to identify genetic modifiers of a cellular process like LDL uptake.
The Scientist's Toolkit: Essential Research Reagents
This table lists key reagents and materials used in the experiments cited, with their primary functions.
| Item | Function / Description |
|---|---|
| Lipid Nanoparticle Spherical Nucleic Acids (LNP-SNAs) | Advanced delivery vehicle that wraps CRISPR machinery in a DNA shell, dramatically improving cellular uptake, editing efficiency, and reducing toxicity [13]. |
| Label IT Tracker Kit (e.g., FITC) | Reagent for covalently labeling plasmid DNA with a fluorophore, enabling tracking of nucleic acid uptake via flow cytometry, independent of gene expression [61] [62]. |
| GeCKOv2 Library | A comprehensive pooled lentiviral library containing ~123,411 gRNAs for genome-scale CRISPR knockout screens in human cells [63]. |
| TransIT-X2 / Lipofectamine 2000 | Cationic polymer/lipid-based transfection reagents used to form complexes with nucleic acids for delivery into a wide range of cell types [61] [58]. |
| Opti-MEM Reduced Serum Medium | A serum-free medium commonly used for diluting DNA and transfection reagents during complex formation to prevent interference from serum components [61] [58]. |
| PureLink HiPure Plasmid Kits | Used for high-quality, high-purity (including endotoxin-free) plasmid midipreps, which are critical for achieving high transfection efficiency and low cell toxicity [61] [58]. |
Troubleshooting Guides
FAQ: How can I design a gRNA to minimize off-target effects?
A: Careful guide RNA (gRNA) design is the most critical step for minimizing off-target effects. A well-designed gRNA maximizes on-target activity while minimizing similarity to other genomic sites. Key strategies include:
- Leverage In-Silico Prediction Tools: Use specialized software to select gRNAs with high predicted on-target activity and low risk for off-target editing. These tools rank potential gRNAs based on sequence uniqueness and other genomic features [64] [65].
- Optimize Sequence Features: Design gRNAs with a GC content between 40% and 60% to stabilize the DNA:RNA duplex at the intended target [66]. The "seed" region (the 12 nucleotides adjacent to the PAM sequence) must be highly specific, with no perfect matches to other genomic sites [67].
- Consider Truncated gRNAs: Using shorter gRNA sequences (17-19 nucleotides instead of 20) can reduce off-target binding without significantly compromising on-target efficiency [68] [66].
- Chemical Modifications: Incorporating chemical modifications like 2'-O-methyl (2'-O-Me) or phosphorothioate (PS) bonds into the gRNA backbone can significantly reduce off-target activity and improve nuclease resistance [64] [66].
FAQ: Which Cas9 variant should I choose for high-specificity editing?
A: Wild-type SpCas9 can tolerate several mismatches, leading to off-target cuts. High-fidelity variants and alternative nucleases have been engineered for greater precision. The choice depends on your specific application and PAM requirements.
Table: High-Fidelity and Alternative Cas Nucleases
| Nuclease | Key Characteristics | Advantages for Reducing Off-Targets | Considerations |
|---|---|---|---|
| SpCas9-HF1 | Engineered high-fidelity variant of SpCas9 [69]. | Greatly reduced off-target cleavage while maintaining robust on-target activity [69]. | On-target efficiency may be reduced in some contexts [64]. |
| eSpCas9 | Engineered high-fidelity variant of SpCas9 [68]. | Designed to minimize non-specific interactions with the DNA backbone. | On-target efficiency may be reduced in some contexts [64]. |
| SuperFi-Cas9 | A redesigned Cas9 from the University of Texas at Austin [66]. | Reported to be 4,000 times less likely to cut at off-target sites [66]. | A recently developed novel variant. |
| SaCas9 | Cas9 from Staphylococcus aureus; requires a different PAM sequence (NNGRRT) [66]. | Its more complex PAM requirement naturally reduces the number of potential off-target sites in the genome [66]. | Larger PAM requirement can limit targetable sites. |
| Cas9 Nickase | A mutated Cas9 that cuts only one DNA strand [67] [66]. | Requires two adjacent gRNAs to create a double-strand break, dramatically increasing specificity [67]. | Requires the design and delivery of two gRNAs. |
| Base Editors | Fuse catalytically impaired Cas9 (dCas9 or nCas9) to a deaminase enzyme [64] [70]. | Do not create double-strand breaks, thereby reducing off-target indels and chromosomal rearrangements [64]. | Potential for guide-independent off-target editing at the RNA or DNA level [70]. |
| Prime Editors | Combine nCas9 with a reverse transcriptase and a prime editing guide RNA (pegRNA) [70] [71]. | Enable precise edits without double-strand breaks, offering high precision and very low off-target rates [70] [71]. | Can be more complex to design and deliver due to the larger size of the editing machinery. |
FAQ: What delivery method is best for reducing off-target effects?
A: The choice of delivery method directly influences how long the CRISPR components are active in the cell, which is a major factor in off-target effects.
- Ribonucleoprotein (RNP) Delivery: This is the preferred method for minimizing off-targets. Directly delivering preassembled Cas9-gRNA complexes results in rapid editing and rapid degradation of the components within the cell. This short activity window (1-2 day half-life) leaves little time for off-target cleavage [3] [66].
- Plasmid or Viral Vector Delivery: These methods result in prolonged expression of Cas9 and gRNA inside the cell, which significantly increases the chance of off-target activity. If using these methods, consider high-fidelity Cas variants and inducible systems to control the duration of expression [64] [68].
FAQ: How do I detect and validate off-target effects in my experiments?
A: After performing gene editing, it is crucial to assess off-target activity, especially for therapeutic applications. Regulatory agencies like the FDA now expect this characterization [64].
- Candidate Site Sequencing: This common method involves sequencing the top potential off-target sites predicted in silico during gRNA design. It is effective if the number of candidate sites is low [64].
- Biased vs. Unbiased Detection Methods:
- Biased Methods: Rely on in silico predictions to identify sites for validation. Examples include tools like Cas-OFFinder and CFD scoring [68].
- Unbiased Methods: Use experimental approaches to genome-widely identify off-target sites without prior prediction. These are more comprehensive and include:
- Whole Genome Sequencing (WGS): WGS is the most comprehensive method to analyze all types of unintended edits, including large chromosomal rearrangements, though it is more expensive and data-intensive [64].
The following diagram illustrates the logical workflow for designing a CRISPR experiment with minimal off-target effects, incorporating the key strategies discussed above.
The Scientist's Toolkit: Essential Reagents for High-Fidelity Editing
Table: Key Research Reagents and Their Functions
| Reagent / Tool | Function / Description | Key Benefit |
|---|---|---|
| Synthetic, Chemically Modified gRNA | gRNAs synthesized with modifications (e.g., 2'-O-Me, PS) to improve stability and specificity [64]. | Reduces off-target editing and increases on-target efficiency. |
| High-Fidelity Cas9 Protein | Purified engineered Cas9 proteins (e.g., SpCas9-HF1, eSpCas9) for RNP complex formation [64] [69]. | Dramatically lower off-target cleavage activity compared to wild-type SpCas9. |
| Cas9 Nickase | A mutated Cas9 that makes single-strand breaks (nicks) instead of double-strand breaks [67] [66]. | Paired nickase strategy requires two adjacent gRNAs for a functional edit, vastly improving specificity. |
| CRISPR Prediction Software (e.g., CRISPOR, DeepCRISPR) | Algorithmic tools to design and rank gRNAs based on on-target efficiency and off-target potential [64] [70]. | Enables data-driven selection of the best gRNA before starting wet-lab experiments. |
| Off-Target Detection Kits (e.g., GUIDE-seq, CIRCLE-seq) | Commercial kits or established protocols for genome-wide identification of off-target sites [64] [68]. | Provides comprehensive experimental data to validate editing specificity and support regulatory filings. |
| Analysis Software (e.g., ICE Tool) | Tools for analyzing Sanger or NGS data to determine editing efficiency and identify off-targets [64]. | Offers fast, robust analysis of editing outcomes for publication-quality data. |
Technical Support Center
Troubleshooting Guides and FAQs
Why does my CRISPR editing in neurons show low efficiency, and how can I improve it?
Low editing efficiency in neurons is often not due to delivery failure but is a consequence of their unique, slow DNA repair processes. Neurons and other postmitotic cells require different optimization strategies than dividing cells.
- Problem Explanation: Unlike dividing cells, which can rapidly repair Cas9-induced double-strand breaks (DSBs) within days, postmitotic human neurons can take several weeks to fully resolve this damage [72]. Furthermore, neurons predominantly employ non-homologous end joining (NHEJ) repair pathways, leading to a different distribution of editing outcomes (smaller indels) compared to the larger deletions often seen in dividing cells [72].
- Solution:
- Extend Your Assay Timeline: When working with neurons, plan your efficiency analysis for up to 16 days or longer post-transduction, as indels continue to accumulate over this period [72].
- Validate Delivery Success: Use a positive control, such as an Adenine Base Editor (ABE), which does not rely on DSB repair. Comparable efficiency between neurons and dividing cells with ABE confirms successful delivery and highlights the DSB repair-specific nature of the delay [72].
- Choose Appropriate gRNAs: Select guide RNAs known to produce efficient NHEJ-like outcomes in non-dividing cells [72].
How can I reduce off-target effects in my CRISPR experiments?
Off-target effects remain a significant hurdle for all cell types. A multi-pronged approach is recommended.
- Problem Explanation: Cas9 can cleave DNA at sites with sequence similarity to the intended target, potentially leading to unintended mutations and confounding experimental results [31].
- Solution:
- Rational gRNA Design and Modification: Use computational tools (e.g., CRISPR-P, E-CRISP) to select highly specific target sequences [73]. Truncating the 5' end of the sgRNA by 2-3 nucleotides can increase binding stringency and reduce off-target cleavage without compromising on-target activity [73].
- Utilize High-Fidelity Cas Variants: Engineered Cas9 proteins like eSpCas9, SpCas9-HF1, and HiFi Cas9 have been designed with mutations that reduce off-target activity while maintaining robust on-target cleavage [31] [73].
- Employ Unbiased Detection Methods: Move beyond computationally predicted off-target screening. Methods like GUIDE-seq and Digenome-seq allow for genome-wide, unbiased identification of DSBs to fully assess editing specificity [31].
My genome editing efficiency seems low across multiple cell types. What general steps can I take?
- Problem Explanation: Low efficiency can stem from various factors, including suboptimal gRNA design, reagent delivery, or cellular health [74].
- Solution:
- Test Multiple Guides: Not all guides targeting a gene perform equally. Design and test 2-3 different gRNAs targeting different PAM sites within your gene of interest to find the most effective one [74].
- Optimize Delivery and Conditions: Use high-quality plasmid DNA or ribonucleoprotein (RNP) complexes [30]. For transfection, optimize conditions such as cell density, reagent concentration, and duration. Adding reagents like DMSO to sequencing reactions can improve results [30].
- Enrich for Edited Cells: Techniques like adding antibiotic selection or using fluorescence-activated cell sorting (FACS) to enrich for successfully transfected cells can dramatically increase the apparent editing efficiency in your population [30].
- Include Rigorous Controls: Always use positive controls (e.g., sgRNAs targeting essential genes like PLK1 or safe harbor loci like AAVS1) and negative controls (non-targeting sgRNAs) to benchmark performance and distinguish specific effects from background noise [75].
Experimental Protocols & Data Analysis
Quantitative Analysis of Cell-Type-Specific Editing Outcomes
The table below summarizes key differences in DNA repair dynamics between dividing cells and postmitotic neurons, which directly impact experimental design and interpretation [72].
| Cell Type | DSB Repair Timeline | Predominant Repair Pathway | Common Indel Spectrum | Key Experimental Consideration |
|---|---|---|---|---|
| Dividing Cells (e.g., iPSCs) | Plateaus within a few days [72] | More MMEJ [72] | Broader range, larger deletions [72] | Standard short-term assays are sufficient. |
| Postmitotic Cells (e.g., Neurons) | Increases for at least 16 days [72] | Predominantly NHEJ [72] | Narrower distribution, smaller indels [72] | Requires long-term assays (2+ weeks). |
Protocol: Analyzing High-Throughput CRISPR Editing Data with CRISPRMatch
For accurate quantification of editing efficiency (indel frequency) from next-generation sequencing (NGS) data, automated pipelines are essential.
- Data Preprocessing: Join paired-end sequencing reads using FLASH to create single, long reads [76] [44].
- Read Mapping: Map the joined reads to the target genomic region using BWA (Burrows-Wheeler Aligner) with default parameters [76] [44].
- Mutation Calling and Classification:
- The pipeline defines the target region for analysis (for SpCas9: 10 bp upstream of gRNA + gRNA + PAM + 10 bp downstream) [76] [44].
- Using Pysam, it detects insertions and deletions in each mapped read.
- Reads are classified into three mutation types: "deletion only," "insertion only," or "both deletion and insertion" [76] [44].
- Efficiency Calculation and Visualization:
- Mapped read counts are normalized (e.g., to one million) to calculate mutation frequency [76] [44].
- The pipeline outputs publication-ready figures, including alignment matrices and plots of deletion frequency at each nucleotide position, allowing for clear evaluation of editing efficiency and accuracy [76] [44].
CRISPRMatch NGS Analysis Workflow
Pathway Diagram: Differential DSB Repair in Dividing vs. Postmitotic Cells
The following diagram illustrates the fundamental differences in how dividing cells and neurons process and repair Cas9-induced DNA breaks, leading to distinct experimental outcomes.
DSB Repair Pathway Differences
The Scientist's Toolkit: Essential Research Reagents
This table lists key reagents and their functions for troubleshooting CRISPR experiments in challenging cell types.
| Reagent / Tool | Function | Application Example |
|---|---|---|
| Virus-Like Particles (VLPs) [72] | Efficient delivery of Cas9 RNP into hard-to-transfect cells (e.g., neurons). | Pseudotyped with VSVG/BRL for high transduction efficiency in human iPSC-derived neurons [72]. |
| High-Fidelity Cas Variants (eSpCas9, SpCas9-HF1) [31] [73] | Engineered Cas9 proteins with reduced off-target effects. | Used in place of wild-type SpCas9 to minimize off-target cleavage in sensitive applications [73]. |
| Positive Control gRNAs (e.g., AAVS1, PLK1) [75] | Benchmark editing efficiency and optimize delivery protocols. | AAVS1 targets a safe harbor locus for neutral edits. PLK1 knockout induces rapid cell death, providing a clear phenotypic readout [75]. |
| Negative Control gRNAs [75] | Non-targeting guides to control for non-specific effects. | Essential in screens and experiments to distinguish true editing effects from background biological noise [75]. |
| CRISPRMatch Software [76] [44] | Automated pipeline for calculating mutation frequency from NGS data. | Analyze batch samples from CRISPR-Cas9 or -Cpf1 experiments to automatically generate efficiency statistics and visualizations [76]. |
| GeneArt Genomic Cleavage Detection Kit [30] | PCR-based kit to verify nuclease cleavage on endogenous genomic DNA. | Used to confirm that low efficiency is not due to an inability of the nuclease to access or cleave the target site [30]. |
Measuring Success: Rigorous Validation Frameworks and Platform Comparisons
The success of CRISPR gene editing experiments hinges on accurate and comprehensive analysis of editing outcomes. Researchers have multiple methodological approaches at their disposal, each with distinct advantages, limitations, and optimal use cases. This technical support center provides troubleshooting guidance and detailed protocols for the most widely used CRISPR analysis techniques, from accessible Sanger sequencing methods to sophisticated high-throughput next-generation sequencing (NGS) approaches. Proper selection and implementation of these analysis methods directly impacts the optimization of CRISPR editing efficiency by providing reliable data for experimental refinement.
Analysis Methodologies and Technical Protocols
ICE (Inference of CRISPR Edits) Analysis
Experimental Protocol for ICE Analysis:
- Sample Preparation: Extract genomic DNA from both edited and unedited (control) cell populations. Amplify the target region using PCR with primers that flank the edit site by at least 200 base pairs on each side [77].
- Sequencing: Submit PCR products for Sanger sequencing according to your institution's core facility guidelines.
- Data Analysis: Upload the Sanger sequencing trace files (.ab1) to the ICE web platform (ice.synthego.com). Input your guide RNA sequence (without PAM) and select the appropriate nuclease (SpCas9, hfCas12Max, Cas12a, or MAD7 are supported) [78].
- Interpretation: Review the ICE Score (editing efficiency percentage), R² value (quality metric), and Knockout Score (proportion of cells with frameshift or 21+ bp indels) [79]. Examine the Contributions, Indel Distribution, and Traces tabs for detailed characterization of editing outcomes.
Troubleshooting FAQs:
- Q: What does a yellow check mark indicate in my ICE results? A: A yellow check mark typically indicates that a parameter was automatically adjusted during analysis. Hover over the symbol for specific details, but results are generally still reliable [79].
Q: How can I improve low R² values in ICE analysis? A: Low R² values suggest poor model fit. Ensure high-quality Sanger sequencing traces, verify your control sample is truly unedited, and confirm correct gRNA sequence input [78].
Q: Can ICE analyze experiments with multiple gRNAs? A: Yes, ICE can analyze multiplexed editing experiments. The platform will visualize all detected edit types and indicate which gRNA was involved in each edit [79].
TIDE (Tracking of Indels by Decomposition) Analysis
Experimental Protocol for TIDE Analysis:
- Follow the same sample preparation and sequencing steps as for ICE analysis [77].
- Upload both edited and control trace files to the TIDE web tool along with your sgRNA sequence.
- The algorithm will decompose the sequence traces and quantify insertion and deletion frequencies within a defined genomic window.
- Review the graphical output representing all indels identified and the estimated editing efficiency of your CRISPR system.
Troubleshooting FAQs:
- Q: What amplification strategy works best for TIDE analysis? A: Design PCR primers to generate an amplicon with at least 200 base pairs of sequence flanking the edit site on either side. This ensures sufficient sequence context for accurate decomposition analysis [77].
- Q: How does TIDE differ from ICE for knockout validation? A: Both methods use Sanger sequencing traces, but TIDE specializes in quantifying indel frequencies while ICE provides additional metrics like Knockout Score that estimates functional knockout likelihood [77] [79].
NGS-Based Analysis Methods
Experimental Protocol for NGS-Based CRISPR Analysis:
- Library Preparation: Amplify target regions from genomic DNA using primers with appropriate adapters for your sequencing platform. For high-throughput screens, this may involve pooled sgRNA library amplification [80].
- Sequencing: Sequence on an appropriate NGS platform (MiSeq for targeted validation; HiSeq for genome-wide screens) to achieve sufficient coverage (≥1000 reads per sample recommended) [81].
- Data Processing:
- Quality Control: Assess sequencing quality metrics, mapping rates, sample correlations, and negative control performance [80].
Troubleshooting FAQs:
- Q: What NGS coverage is sufficient for CRISPR validation studies? A: GENEWIZ's genoTYPER-NEXT assay typically provides at least 1,000 paired reads per sample, which is sufficient for most validation studies [81]. For quantitative applications, higher coverage may be necessary.
Q: How can I assess the quality of my CRISPR screen data? A: MAGeCK-VISPR provides multi-level QC metrics: sequence level (GC content, base quality), read count level (mapping rates, Gini index), sample level (correlations, PCA), and gene level (ribosomal gene depletion) [80].
Q: What if my target region is larger than standard amplicon limits? A: Custom assays can be designed to cover larger target regions. Discuss with sequencing service providers for tailored solutions [81].
Comparative Analysis of CRISPR Methods
Table 1: Comparison of Major CRISPR Analysis Methods
| Method | Throughput | Cost Factor | Primary Applications | Key Metrics | Advantages | Limitations |
|---|---|---|---|---|---|---|
| ICE | Medium | 1x | Knockout validation, Efficiency assessment | ICE Score, R², KO Score | ~100x cost reduction vs NGS, Quantitative from Sanger data [78] | Limited to edits near cut site |
| TIDE | Medium | 1x | Indel frequency quantification | Indel spectrum, Editing efficiency | Simple workflow, No special software required [77] | Less accurate for complex edits |
| NGS-Targeted | High | ~100x | Comprehensive edit characterization, Off-target assessment | Insertion/deletion sequences, Editing efficiency | Base-resolution detail, Detects complex edits [44] [77] | Higher cost, Computational requirements |
| NGS-Pooled Screens | Very High | Varies | Functional genomics, Gene essentiality | Gene essentiality scores, sgRNA abundance | Genome-wide coverage, Phenotypic coupling [80] | Complex analysis, Specialized libraries required |
Table 2: Computational Tools for CRISPR Analysis
| Tool | Platform | Input Data | Key Features | Supported Nucleases |
|---|---|---|---|---|
| CRISPRMatch | Stand-alone (Python) | NGS data | Automatic calculation, Visualization, Batch processing | Cas9, Cpf1 [44] |
| MAGeCK-VISPR | Stand-alone | NGS screens | Quality control, Multi-condition analysis, Visualization | Cas9, dCas9 [80] |
| ICE | Web-based | Sanger traces | Knockout score, No software installation | SpCas9, hfCas12Max, Cas12a, MAD7 [78] |
| TIDE | Web-based | Sanger traces | Indel decomposition, No specialized equipment | Cas9 [77] |
Research Reagent Solutions
Table 3: Essential Reagents for CRISPR Analysis Workflows
| Reagent/Cell Line | Function | Application Examples |
|---|---|---|
| Modified Guide RNAs | Enhanced stability and reduced immune response; chemical modifications like 2'-O-methyl at terminal residues improve editing efficiency [82] | Genome editing in primary cells and hard-to-transfect cell types |
| Ribonucleoproteins (RNPs) | Cas protein pre-complexed with guide RNA; increases editing efficiency, reduces off-target effects, enables DNA-free editing [82] | Therapeutic applications, sensitive cell types |
| Stably Expressing Cas9 Cell Lines | Consistent Cas9 expression; improves reproducibility, eliminates transfection variability [83] | High-throughput screens, recurrent editing experiments |
| hiNLS Cas9 Variants | Enhanced nuclear localization via hairpin internal nuclear localization signals; improves editing efficiency in primary cells [3] | Therapeutic applications in primary human lymphocytes |
| Selectable Markers | Antibiotic resistance or fluorescent reporters; enables enrichment of successfully transfected cells [38] | Isolation of edited populations, tracking transduction efficiency |
Workflow Visualization
CRISPR Analysis Method Selection Workflow
Advanced Troubleshooting Guide
Addressing Low Editing Efficiency
Q: My CRISPR experiments consistently show low editing efficiency across validation methods. What optimization strategies should I prioritize?
A: Low editing efficiency can stem from multiple factors. Implement these evidence-based solutions:
- sgRNA Optimization: Test 2-3 different guide RNAs targeting the same gene to identify the most effective sequence. Bioinformatics tools can predict optimal guides, but empirical testing remains essential [82].
- Delivery Method Enhancement: For hard-to-transfect cells, switch to electroporation or viral delivery instead of lipid-based methods. Consider using ribonucleoprotein (RNP) complexes rather than plasmid DNA, as RNPs can increase editing efficiency and reduce off-target effects [82] [83].
- Nuclear Localization Improvement: Utilize Cas9 variants with enhanced nuclear localization signals (e.g., hiNLS constructs) to improve nuclear import, particularly important for primary cells and therapeutic applications [3].
- Verification of Component Concentration: Precisely quantify guide RNA concentrations and maintain appropriate guide:nuclease ratios to maximize editing while minimizing cellular toxicity [82].
Analyzing Complex Editing Outcomes
Q: How can I accurately characterize samples edited with multiple gRNAs or containing complex indel patterns?
A: Complex editing scenarios require advanced approaches:
- Multiplexed gRNA Experiments: Use ICE analysis for Sanger-based characterization of samples with multiple gRNAs. The platform identifies which gRNA generated each edit and provides visualizations of complex outcomes [79].
- NGS for Complex Patterns: For highly heterogeneous editing or large deletions, employ targeted NGS with tools like CRISPRMatch, which automatically classifies different mutation types (deletions, insertions, or both) and provides visualization of editing patterns across the target region [44].
- Experimental Design Simplification: When possible, use single gRNA approaches or sequential editing to reduce complexity. For large deletions, design dual gRNAs flanking the region of interest and screen by PCR size selection before detailed characterization [77].
Selecting appropriate analysis methods is crucial for accurate interpretation of CRISPR editing outcomes and meaningful optimization of editing efficiency. The method selection should be guided by experimental goals, resource constraints, and required resolution. Sanger-based methods (ICE, TIDE) offer cost-effective solutions for routine validation, while NGS approaches provide comprehensive characterization for complex edits and discovery applications. As CRISPR technologies evolve toward therapeutic applications, robust analysis methods will play an increasingly critical role in ensuring both efficacy and safety of genome editing interventions.
Frequently Asked Questions (FAQs)
Q1: Why is the choice of DNA repair pathway different in dividing versus non-dividing cells? The fundamental difference lies in the availability of a sister chromatid to use as a repair template. Dividing cells, particularly those in the S and G2 phases of the cell cycle, primarily use the high-fidelity Homologous Recombination (HR) pathway because a homologous template is present. Non-dividing cells, which are in the G0 or G1 phase, lack this readily available template and therefore rely almost exclusively on the faster but more error-prone Non-Homologous End Joining (NHEJ) pathway to repair DNA double-strand breaks [84] [85]. This has direct implications for CRISPR editing outcomes, as NHEJ typically results in small insertions or deletions (indels), while HR can enable precise gene insertion or correction.
Q2: How does the cellular division state influence the efficiency of CRISPR-HDR editing? Homology-Directed Repair (HDR) is much more efficient in dividing cells because it is tightly coupled with the cell cycle and depends on proteins that are active during the S and G2 phases [84]. In non-dividing or slowly dividing cells, HDR efficiency is notoriously low because the necessary machinery is less active and the preferred NHEJ pathway dominates. For projects requiring precise HDR in non-dividing cells (e.g., neuronal or primary cell editing), consider using alternative editors like prime editing or base editing that can operate more independently of the cell cycle and do not require a donor template [38].
Q3: What are the practical consequences of NHEJ-dominated repair in non-dividing cells for my gene knockout experiment? In non-dividing cells, the prevalence of NHEJ is advantageous for generating gene knockouts, as it efficiently creates disruptive indels at the target site [38]. However, the heterogeneous mixture of indels can lead to a mosaic of knockout efficiencies. The main challenge is that the error-prone nature of NHEJ can sometimes lead to unpredictable editing outcomes or, in the case of large deletions, potential genomic instability. Using validated, highly efficient guide RNAs is critical to maximize the percentage of cells with the desired knockout [86].
Q4: My editing efficiency in primary T cells is low. What strategies can I use to improve it? Low efficiency in primary cells like T lymphocytes is a common challenge. Recent research demonstrates that optimizing the nuclear delivery of CRISPR components can significantly boost efficiency. Strategies include:
- Using Ribonucleoproteins (RNPs): Delivering pre-assembled Cas9-protein and guide RNA complexes reduces toxicity and can increase editing rates [86] [3].
- Engineering Enhanced Nuclear Localization Signals (NLS): Novel constructs, such as hairpin internal NLS (hiNLS) fused to Cas9, have been shown to enhance nuclear import and significantly improve knockout efficiency in primary human T cells compared to standard terminally-fused NLS [3].
- Electroporation: This is a highly effective delivery method for hard-to-transfect primary cells [3].
Troubleshooting Guides
Problem: Low HDR Efficiency in Dividing Cells
Potential Causes and Solutions:
| Cause | Solution |
|---|---|
| Low proportion of cells in S/G2 phase | Synchronize the cell cycle before transfection to enrich for cells in S/G2 phase. |
| NHEJ outcompeting HDR | Use an NHEJ inhibitor (e.g., small molecule compounds like Scr7) during editing to tilt the balance toward HDR [38]. |
| Inefficient delivery of donor template | Optimize the donor DNA design (e.g., use single-stranded oligodeoxynucleotides for small insertions) and ensure it is co-delivered effectively with the CRISPR machinery [38]. |
| Low Cas9 expression/activity | Use a high-activity Cas9 variant and verify protein expression. Consider RNP delivery for more immediate and uniform activity [86]. |
Problem: Inconsistent or Mosaic Editing in Non-Dividing Cell Populations
Potential Causes and Solutions:
| Cause | Solution |
|---|---|
| Variable RNP delivery or Cas9 expression | Use Ribonucleoprotein (RNP) complex delivery via electroporation to ensure all cells receive a similar dose of editing components simultaneously, reducing mosaic outcomes [86]. |
| Slow or delayed Cas9 nuclear import | Employ Cas9 constructs with advanced nuclear localization signals (hiNLS) to accelerate nuclear entry, which is crucial for efficient editing in non-dividing cells before the RNP is degraded [3]. |
| Guide RNA with low intrinsic efficiency | Design and test multiple guide RNAs (typically 3-4) targeting your gene of interest to identify the most effective one. Use bioinformatics tools to aid in selection [86] [67]. |
The table below summarizes the core DNA repair pathways relevant to CRISPR-Cas9 gene editing.
Table 1: Key DNA Double-Strand Break Repair Pathways
| Feature | Non-Homologous End Joining (NHEJ) | Homologous Recombination (HR) |
|---|---|---|
| Primary Cellular Context | Active throughout cell cycle; dominant in non-dividing (G0/G1) cells [84] [85]. | Primarily active in S and G2 phases in dividing cells [84] [85]. |
| Template Required | No | Yes (sister chromatid or homologous chromosome) |
| Fidelity | Error-prone; often results in small insertions or deletions (indels) [38]. | High-fidelity; accurately restores the original DNA sequence [38]. |
| Key Proteins | Ku70/Ku80, DNA-PKcs, XRCC4, DNA Ligase IV [87] [85]. | MRN complex, CtIP, BRCA1, Rad51 [84] [85]. |
| Primary CRISPR Outcome | Gene Knockouts (via frameshift mutations) | Precise Gene Insertion/Correction (via HDR) |
Experimental Protocols
Protocol 1: Enhancing CRISPR-KO in Non-Dividing Cells using RNP Electroporation
This protocol is optimized for achieving high knockout efficiency in hard-to-transfect non-dividing cells, such as primary human T lymphocytes [86] [3].
- Guide RNA Design: Design and procure 2-3 chemically synthesized, modified guide RNAs targeting your gene. Using modified guides improves stability and reduces immune stimulation [86].
- RNP Complex Formation:
- Resuspose Alt-R S.p. Cas9 Nuclease (or a hiNLS-Cas9 variant [3]) in nuclease-free duplex buffer to a concentration of 10 µM.
- Resuspend the lyophilized guide RNA in nuclease-free duplex buffer to a concentration of 100 µM.
- To form the RNP complex, mix the Cas9 nuclease and guide RNA at a molar ratio of 1:2 to 1:5 (e.g., 5 µL of 10 µM Cas9 with 2.5 µL of 100 µM gRNA). Incubate at room temperature for 10-20 minutes.
- Cell Preparation: Isolate and wash your primary cells. Count and resuspend them in the recommended electroporation buffer at a concentration of 1-2 x 10^6 cells per 10 µL.
- Electroporation: Mix 10 µL of cell suspension with the pre-formed RNP complex. Transfer the entire mixture to an electroporation cuvette. Electroporate using a device- and cell-type-specific program (e.g., for T cells, a protocol using the Lonza 4D-Nucleofector system is common).
- Recovery and Analysis: Immediately after electroporation, add pre-warmed culture media to the cells and transfer them to a culture plate. Allow cells to recover for 48-72 hours before analyzing editing efficiency via genomic DNA extraction, PCR, and sequencing (e.g., T7 Endonuclease I assay or NGS) [86].
Protocol 2: Optimizing HDR in Dividing Cells
This protocol outlines steps to maximize HDR efficiency for precise editing in actively dividing cell lines [38].
- Cell Cycle Synchronization: Treat your cell culture with a reagent like thymidine or nocodazole to arrest the majority of cells at the G1/S boundary or in mitosis. Release the arrest just before transfection to enrich the population of cells entering S phase, where HR is most active.
- Donor Template Design: For small insertions (<60 bp), design a single-stranded oligodeoxynucleotide (ssODN) donor with homologous arms (30-50 bp on each side). For larger insertions (up to 5 kb), use a double-stranded DNA (dsDNA) donor, such as a plasmid, with longer homology arms (≥500 bp).
- CRISPR Component Delivery: Co-deliver the Cas9/gRNA (as plasmid, mRNA, or RNP) and the donor template into the synchronized cells using your preferred method (e.g., lipofection, nucleofection).
- HDR Enhancement: After delivery, add a commercially available HDR enhancer solution to the culture media. Alternatively, supplement the media with a small molecule NHEJ inhibitor.
- Isolation and Validation: Allow 48-72 hours for repair. If using a fluorescent or antibiotic selection marker in your donor template, use FACS sorting or antibiotic selection to enrich for successfully edited cells. Validate precise edits via PCR and Sanger sequencing.
Signaling Pathways and Experimental Workflows
DNA Repair Pathway Choice in Response to CRISPR-Induced Breaks
Workflow for Editing Non-Dividing vs. Dividing Cells
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Reagents for Cell-Type Specific CRISPR Editing
| Item | Function | Application Note |
|---|---|---|
| hiNLS-Cas9 Constructs | Engineered Cas9 with enhanced nuclear localization signals for improved nuclear import and editing efficiency. | Critical for enhancing knockout rates in non-dividing primary cells like T lymphocytes [3]. |
| Chemically Modified sgRNA | Synthetic guide RNAs with molecular modifications (e.g., 2'-O-methyl) to increase stability and reduce cellular immune responses. | Recommended over in vitro transcribed (IVT) guides for all cell types, especially sensitive primary cells [86]. |
| Ribonucleoprotein (RNP) Complex | Pre-assembled complex of Cas9 protein and sgRNA. | The preferred delivery form for reducing off-target effects and achieving rapid, highly efficient editing, particularly in non-dividing cells [86] [3]. |
| NHEJ Inhibitors (e.g., Scr7) | Small molecules that temporarily inhibit key proteins in the NHEJ pathway. | Used in dividing cells to shift the repair balance toward the HDR pathway for precise editing [38]. |
| HDR Enhancer Solutions | Commercial reagent formulations designed to improve the efficiency of homology-directed repair. | Added to culture media post-transfection to boost HDR rates in dividing cells [38]. |
| Electroporation Systems | Hardware for delivering macromolecules (like RNPs) into cells via electrical pulses. | Essential for efficient delivery into hard-to-transfect non-dividing and primary cells [3]. |
In CRISPR gene editing, accurately measuring efficiency is fundamental for developing robust therapeutic and research applications. Efficiency metrics quantitatively assess the success of your editing experiment, distinguishing between mere delivery of editing components (transfection) and successful alteration of the target DNA (editing). Standardized measurements for knockout and correction rates allow for the comparative evaluation of different guide RNAs (gRNAs), delivery methods, and experimental conditions, ultimately leading to more reliable and reproducible outcomes in your research [88] [89].
This guide provides troubleshooting advice and standardized methodologies to help you accurately quantify and optimize your CRISPR editing experiments.
Standardized Methods for Measuring Editing Efficiency
Multiple techniques are available for assessing CRISPR efficiency, each with unique strengths, limitations, and appropriate use cases. The table below summarizes the most common methods.
| Method | Key Principle | Typical Data Output | Advantages | Limitations |
|---|---|---|---|---|
| T7 Endonuclease I (T7EI) Assay [88] | Mismatch-specific enzyme cleaves heteroduplex DNA formed by wild-type and indel-containing strands. | Semi-quantitative; gel band intensity ratio (cleaved/uncleaved). | Rapid, low-cost, accessible. | Semi-quantitative, lacks sensitivity for low-efficiency editing, potential for underestimation [88]. |
| Tracking of Indels by Decomposition (TIDE) [88] | Algorithm-based decomposition of Sanger sequencing chromatograms from edited populations. | Quantitative; percentage of indels and their spectra. | More quantitative than T7EI, provides indel sequence information. | Accuracy relies on high-quality PCR and sequencing; can miss complex edits [88]. |
| Inference of CRISPR Edits (ICE) [88] | Similar to TIDE, uses sequencing trace decomposition to infer editing efficiency. | Quantitative; editing efficiency and indel profiles. | More quantitative than T7EI, user-friendly software available. | Similar to TIDE, performance depends on sequencing quality [88]. |
| Droplet Digital PCR (ddPCR) [88] | Uses differentially labeled fluorescent probes to distinguish and absolutely quantify edited vs. wild-type alleles in a water-oil emulsion droplet system. | Highly precise and quantitative; absolute count of edited and unedited molecules. | High precision, absolute quantification, excellent for discriminating between edit types (e.g., NHEJ vs. HDR) [88]. | Requires specific equipment and probe design, less effective for novel or undefined indels. |
| Next-Generation Sequencing (NGS) | High-throughput sequencing of the target locus, providing a comprehensive view of all editing outcomes. | Highly quantitative; precise frequency of every indel and substitution. | Gold standard for comprehensiveness and accuracy; reveals full complexity of editing outcomes. | Higher cost, more complex data analysis and bioinformatics requirements [90]. |
| Fluorescent Reporter Cells [88] | Live-cell system where successful installation of an edit (e.g., HDR) activates a fluorescent protein. | Quantitative via flow cytometry or microscopy; percentage of fluorescent cells. | Allows for live-cell tracking and sorting of edited cells. | Only reports on edits at the reporter locus, not the endogenous target; requires cell engineering [88]. |
Choosing the Right Measurement Method
Selecting an appropriate method depends on your experimental goals and resources:
- For quick, low-cost validation of high-efficiency knockouts, the T7EI assay may be sufficient.
- For detailed, quantitative analysis of indel spectra without NGS, TIDE or ICE are strong choices.
- For high-precision quantification of specific allelic changes (e.g., HDR), ddPCR is ideal.
- For the most comprehensive and accurate analysis, especially in a therapeutic context, NGS is recommended.
- For tracking editing in real-time or for enriching edited cell populations, fluorescent reporters are highly effective.
Frequently Asked Questions (FAQs)
My editing efficiency is low. What are the primary factors I should troubleshoot?
Low editing efficiency is often a multi-factorial problem. The most common areas to investigate are:
- gRNA Quality and Design: Ensure your gRNA has high predicted activity. Use design tools and prioritize gRNAs with higher GC content (e.g., ~65%) [28]. Always test 3-4 different gRNA sequences per target [89].
- Delivery Efficiency: The method used to deliver CRISPR components (RNP, plasmid, mRNA) must be efficient for your specific cell type. What works for one cell line may fail in another [91] [89].
- Component Dosage and Ratio: The amounts and ratio of Cas9 to gRNA are critical. Titrate these components to find the optimal balance between high editing and low toxicity [67].
- Target Sequence Accessibility: The chromatin state of your target locus (open or closed) can significantly impact Cas9 access and efficiency [88].
How can I improve editing efficiency in hard-to-transfect primary cells?
Primary cells, like chondrocytes or immune cells, are often challenging. Key strategies include:
- Use Cas9 Ribonucleoprotein (RNP) Complexes: Electroporation of pre-assembled Cas9-gRNA RNPs is the gold standard for primary cells. It is fast, minimizes off-target effects, and has been shown to achieve >90% knockout efficiency in human dendritic cells and chondrocytes [91] [92].
- Optimize Transfection Parameters Systematically: Don't rely on standard protocols. Perform a grid search of electroporation conditions (voltage, pulse length, etc.). Some platforms test up to 200 conditions to find the optimal setting for a given cell type [89].
- Enhance Nuclear Import: Using Cas9 constructs with advanced nuclear localization signals (NLS), such as hairpin internal NLS (hiNLS), can significantly boost editing efficiency by ensuring more Cas9 reaches the nucleus quickly, which is crucial for transient RNP delivery [3].
What is the difference between transfection efficiency and editing efficiency?
This is a critical distinction:
- Transfection Efficiency: The percentage of cells that have successfully taken up the CRISPR components (e.g., are fluorescent if a fluorescent marker is co-delivered).
- Editing Efficiency: The percentage of cells that contain actual modifications at the target genomic locus.
You can have 100% transfection efficiency but 0% editing efficiency if the components are not functional or cannot access the nucleus. Always measure editing efficiency directly via genotyping (e.g., T7EI, TIDE, NGS) rather than relying on transfection markers [89].
How do I accurately measure efficiency when using multiple gRNAs?
When using several gRNAs to target one gene or to create large deletions, standard methods like TIDE might not capture the full spectrum of edits. In these cases:
- Next-Generation Sequencing (NGS) is the most reliable method as it can identify and quantify complex mixtures of indels and large deletions.
- Utilize analysis pipelines that can account for amplicon size biases during PCR, which is particularly important when quantifying large deletions [92].
My positive control works, but my experimental gRNA does not. What should I do?
If a validated positive control gRNA edits efficiently in your system, but your target-specific gRNA does not, the issue is likely with the experimental gRNA itself or the target site.
- Re-evaluate gRNA Design: Re-run the sequence through prediction tools. Check for potential secondary structures in the gRNA or target DNA.
- Check for "Bad Seeds": Ensure the gRNA sequence does not contain problematic motifs, such as a string of T's (TTTT) that can act as a premature termination signal for U6 polymerase [93].
- Test Additional gRNAs: Always design and test multiple (3-4) gRNAs against different regions of your target gene [89].
The Scientist's Toolkit: Essential Reagents & Materials
| Reagent / Material | Function / Explanation |
|---|---|
| Cas9 Nuclease (Wild-type) | The "molecular scissors" that creates double-strand breaks in DNA at the location specified by the gRNA. |
| Synthetic sgRNA | A single guide RNA that combines the functions of crRNA and tracrRNA, directing Cas9 to the specific target genomic sequence. |
| Ribonucleoprotein (RNP) Complex | The pre-assembled complex of Cas9 protein and sgRNA. Direct RNP delivery is highly efficient and reduces off-target effects. |
| Electroporator / Nucleofector | Instrument for delivering CRISPR components (especially RNPs) into cells via electrical pulses, essential for hard-to-transfect cells. |
| Lipid Nanoparticles (LNPs) | An alternative non-viral delivery vehicle for encapsulating and delivering CRISPR components into cells. |
| T7 Endonuclease I | Enzyme used in the T7EI assay to detect and cleave mismatched DNA heteroduplexes, revealing the presence of indels. |
| Droplet Digital PCR (ddPCR) System | Platform for absolute quantification of editing efficiency using a water-oil emulsion droplet system and probe-based detection. |
| Positive Control gRNA | A validated gRNA targeting a well-characterized locus (e.g., AAVS1). Crucial for optimizing delivery parameters and confirming system functionality [89]. |
| Fluorescent Reporter Cell Line | Engineered cells that express a fluorescent protein upon successful gene editing, enabling rapid assessment and sorting of edited populations [88]. |
Detailed Experimental Protocols
Protocol 1: High-Efficiency RNP Electroporation in Primary Human Cells
This protocol is adapted from successful knockout studies in primary human chondrocytes and dendritic cells, achieving efficiencies of ~90% or higher [91] [92].
Workflow Diagram: RNP Electroporation for Primary Cells
Materials:
- Primary cells (e.g., monocytes, chondrocytes)
- Recombinant Cas9 protein
- Synthetic sgRNA (targeting your gene of interest)
- Electroporation system (e.g., Lonza 4D-Nucleofector)
- Appropriate electroporation kit (e.g., P3 Primary Cell Kit)
Step-by-Step Method:
- Cell Preparation: Differentiate your primary cells as required. For monocyte-derived dendritic cells (moDCs), isolate CD14+ monocytes and culture with GM-CSF and IL-4 for 5-7 days [92].
- RNP Complex Assembly: Resuspend synthetic sgRNA in nuclease-free buffer. Mix with Cas9 protein to form the RNP complex. A typical ratio is 1:2 (Cas9:sgRNA, mol/mol). Incubate at room temperature for 10-20 minutes to allow complex formation.
- Cell Harvest: Collect the cells and centrifuge. Aspirate the supernatant completely.
- Cell Resuspension: Resuspend the cell pellet in the provided electroporation solution. Use 1x10^6 to 2x10^6 cells per 20 µL reaction.
- Electroporation: Add the pre-assembled RNP complex to the cell suspension. Transfer the entire mixture to a certified cuvette. Electroporate using the optimized program (e.g., "DJ-108" for moDCs [92] or "EO-115" for chondrocytes [91]).
- Cell Recovery: Immediately after electroporation, add pre-warmed culture medium to the cuvette and transfer the cells to a culture plate.
- Incubation and Analysis: Culture the cells for 48-72 hours to allow for expression of the knockout phenotype. Harvest cells and extract genomic DNA for efficiency analysis via your chosen method (e.g., NGS, TIDE).
Protocol 2: Quantitative Efficiency Analysis using Droplet Digital PCR (ddPCR)
ddPCR provides absolute quantification of editing rates without the need for standard curves, making it highly precise for measuring NHEJ or HDR frequencies [88].
Workflow Diagram: ddPCR for Editing Efficiency
Materials:
- Genomic DNA from edited and control cells
- ddPCR Supermix for Probes (no dUTP)
- FAM-labeled probe for the edited allele
- HEX- or VIC-labeled probe for the wild-type allele
- Droplet generator and reader
Step-by-Step Method:
- DNA Preparation: Extract high-quality genomic DNA. Digest the DNA with a restriction enzyme that does not cut within the amplicon to reduce viscosity and improve droplet generation, if necessary.
- Reaction Setup: Prepare a PCR reaction mix containing ddPCR supermix, forward and reverse primers that flank the edit site, and the two fluorescent probes (FAM for edit, HEX for wild-type).
- Droplet Generation: Transfer the reaction mix to a droplet generation cartridge. Following the manufacturer's instructions, generate thousands of nanoliter-sized water-in-oil droplets.
- PCR Amplification: Transfer the droplets to a 96-well PCR plate. Perform PCR amplification in a thermal cycler using standard cycling conditions optimized for your assay.
- Droplet Reading: Place the plate in the droplet reader. The instrument streams each droplet past a two-color optical detection system, classifying each as FAM-positive (edited), HEX-positive (wild-type), both (heterozygous), or negative (no template).
- Data Analysis: Use the instrument's software (e.g., QuantaSoft) to calculate the absolute concentration (copies/µL) of edited and wild-type alleles. Editing efficiency is calculated as:
[FAM-positive] / ([FAM-positive] + [HEX-positive]) * 100.
Genome editing technologies have revolutionized biological research and therapeutic development by enabling precise modifications to DNA. Among the most powerful tools in this field are Zinc Finger Nucleases (ZFNs), Transcription Activator-Like Effector Nucleases (TALENs), and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) systems. These technologies function as molecular scissors, creating targeted double-strand breaks in DNA that stimulate the cell's natural repair mechanisms. The subsequent repair processes allow researchers to disrupt gene function, correct mutations, or insert new genetic sequences. This technical support center provides a comprehensive benchmarking analysis of these three platforms, focusing on their mechanistic principles, comparative performance, and practical troubleshooting for research applications. With the global genome editing market projected to grow from $10.8 billion in 2025 to $23.7 billion by 2030, representing a compound annual growth rate of 16.9%, mastery of these technologies has become increasingly essential for researchers and drug development professionals [94].
Fundamental Mechanisms of Action
All three genome editing platforms function by creating targeted double-strand breaks (DSBs) in DNA, which activate cellular repair mechanisms. The non-homologous end joining (NHEJ) pathway often results in insertions or deletions (indels) that disrupt gene function, while homology-directed repair (HDR) can introduce precise genetic modifications using a donor DNA template [95] [96]. Despite this shared outcome, each technology employs distinct molecular mechanisms for DNA recognition and cleavage, leading to significant differences in specificity, efficiency, and practical implementation.
Technology Comparison Table
Table 1: Comparative analysis of ZFN, TALEN, and CRISPR genome editing platforms
| Feature | ZFNs | TALENs | CRISPR-Cas9 |
|---|---|---|---|
| DNA Recognition Mechanism | Protein-DNA (Zinc finger domains) | Protein-DNA (TALE repeats) | RNA-DNA (guide RNA) |
| Recognition Pattern | 3 bp per zinc finger domain | 1 bp per TALE repeat | 20 nt guide RNA sequence + PAM |
| Typical Target Length | 9-18 bp | 14-20 bp | 20 bp + NGG PAM |
| Nuclease Domain | FokI (requires dimerization) | FokI (requires dimerization) | Cas9 (functions as monomer) |
| Specificity Mechanism | Dual recognition sites with spacer | Dual recognition sites with spacer | Single guide RNA with PAM constraint |
| Engineering Approach | Complex protein engineering | Modular protein assembly | Simple RNA design |
| Multiplexing Capability | Limited | Limited | High (multiple gRNAs) |
| Primary Advantage | Established technology, smaller size | High specificity, simple code | Easy design, high efficiency, multiplexing |
| Primary Limitation | Context-dependent effects, difficult design | Large size, repetitive sequences | PAM requirement, off-target effects |
Molecular Architecture
Zinc Finger Nucleases (ZFNs) are fusion proteins comprising an engineered zinc finger DNA-binding domain fused to the FokI nuclease domain. Each zinc finger recognizes approximately 3 base pairs, and arrays are typically designed with 3-6 fingers to achieve specificity for 9-18 base pair sequences. A significant constraint is that ZFNs must function as heterodimers, with two separate ZFN pairs binding to opposite DNA strands in a precise orientation and spacing (typically 5-7 bp apart) for FokI dimerization and subsequent DNA cleavage [97] [96].
Transcription Activator-Like Effector Nucleases (TALENs) similarly fuse a DNA-binding domain to the FokI nuclease. However, TALENs utilize TALE repeats derived from Xanthomonas bacteria, with each repeat comprising 33-35 amino acids that recognize a single DNA base pair through two hypervariable residues known as Repeat Variable Diresidues (RVDs). The simple recognition code (NI = A, NG = T, HD = C, NN = G/K) enables more straightforward design compared to ZFNs. Like ZFNs, TALENs function as pairs binding to opposite DNA strands with a spacer sequence between them [97] [96].
CRISPR-Cas9 systems operate through a fundamentally different mechanism involving RNA-DNA recognition rather than protein-DNA interactions. The core components include the Cas9 nuclease and a single guide RNA (sgRNA) that combines the functions of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA). The sgRNA directs Cas9 to complementary DNA sequences adjacent to a Protospacer Adjacent Motif (PAM), which for the most commonly used Streptococcus pyogenes Cas9 is 5'-NGG-3'. Cas9 then induces a double-strand break approximately 3-4 bp upstream of the PAM sequence [95] [97].
Troubleshooting Guide: Common Experimental Challenges
Low Editing Efficiency
Problem: Inadequate modification at the target locus across all platforms.
Solutions:
- CRISPR-Specific Optimization:
- Design and test 3-4 different guide RNAs targeting the same locus to identify the most effective sequence [67].
- Increase the length of tracrRNA, as consistent increases in modification efficiency have been observed with longer tracrRNA variants [67].
- Utilize AI-based gRNA design tools (e.g., DeepCRISPR, CRISPRon) that predict high-activity guide RNAs using machine learning models trained on large experimental datasets [70].
- Employ advanced delivery systems such as Lipid Nanoparticle Spherical Nucleic Acids (LNP-SNAs), which have demonstrated threefold improvements in editing efficiency compared to standard delivery methods [13].
Platform Selection Considerations:
- If using CRISPR, verify PAM availability and consider Cas9 variants with alternate PAM specificities if the target region lacks conventional NGG sites [95].
- For TALENs, ensure proper spacer length between binding sites (typically 12-20 bp) to allow FokI dimerization [97].
- With ZFNs, verify that binding sites are appropriately oriented and spaced for functional dimerization [96].
General Optimization Strategies:
- Enrich transfected cells through antibiotic selection or FACS sorting to increase the population of successfully modified cells [67].
- Optimize delivery method and timing for specific cell types, as efficiency varies significantly across different cell lines and primary cells [9].
- Validate component expression and quality, particularly when using plasmid-based systems where promoter compatibility or DNA degradation can impact results [9].
Off-Target Effects
Problem: Unintended modifications at genomic sites with similarity to the target sequence.
Solutions:
- CRISPR-Specific Approaches:
- Titrate sgRNA and Cas9 concentrations to optimize the on-target to off-target cleavage ratio, as reduced concentrations can improve specificity while potentially maintaining on-target activity [67].
- Utilize high-fidelity Cas9 variants (e.g., eSpCas9, SpCas9-HF1) engineered to reduce off-target cleavage while maintaining on-target efficiency [95].
- Employ Cas9 nickase mutants that create single-strand breaks rather than double-strand breaks, requiring paired guide RNAs for effective cleavage, thereby dramatically increasing specificity [67].
- Design guide RNAs with highly unique 12-nucleotide "seed" sequences adjacent to the PAM and ensure at least two mismatches in the PAM-proximal region for any potential off-target sites [67].
- Leverage bioinformatic tools (e.g., Cas-OFFinder, CCTop) to predict and evaluate potential off-target sites during guide RNA design [98].
Platform Selection Considerations:
- TALENs inherently exhibit higher specificity due to the requirement for dimerization and longer recognition sequences, making them preferable for applications where utmost precision is critical [97].
- Consider the structural constraints of ZFNs, where the necessity for dual binding sites provides inherent specificity safeguards [96].
Validation Methods:
- Employ genome-wide off-target detection techniques such as GUIDE-seq, CIRCLE-seq, or DISCOVER-Seq to empirically identify unintended editing events [98].
- Implement rigorous molecular screening including T7 endonuclease I assays, Surveyor assays, or comprehensive sequencing to validate editing specificity [9] [67].
Cytotoxicity and Cell Viability Issues
Problem: Reduced cell survival following editing component delivery.
Solutions:
- Delivery Optimization:
- Utilize advanced delivery systems such as LNP-SNAs, which have demonstrated significantly reduced toxicity compared to standard lipid nanoparticles or viral vectors [13].
- Optimize the concentration of editing components, starting with lower doses and titrating upward to identify the balance between editing efficiency and cell viability [9].
- Consider ribonucleoprotein (RNP) delivery of precomplexed Cas9 and sgRNA rather than plasmid DNA, which can reduce prolonged nuclease expression and associated toxicity [9].
Platform Considerations:
Experimental Design:
- Include appropriate controls to distinguish between platform-specific toxicity and general transfection-related stress.
- Monitor timing of expression and consider inducible systems to limit prolonged nuclease activity [9].
PAM Sequence Limitations
Problem: Lack of suitable PAM sequences adjacent to the target site for CRISPR systems.
Solutions:
- Alternative PAM Recognition:
- For SpCas9, consider NAG as an alternative PAM sequence, though with approximately one-fifth the efficiency of canonical NGG PAMs [67].
- Explore engineered Cas9 variants with altered PAM specificities (e.g., xCas9, SpCas9-NG) that recognize broader PAM sequences [95].
- Utilize orthologous Cas proteins from different bacterial species (e.g., St1Cas9, Cpf1) with distinct PAM requirements [65].
- Platform Alternatives:
Detection and Validation Challenges
Problem: Difficulty in confirming successful edits, particularly in mixed cell populations.
Solutions:
- Screening Method Selection:
- Employ PCR-based screening followed by restriction fragment length polymorphism (RFLP) analysis using enzymes like T7 endonuclease I or Surveyor nuclease to detect indels in heterogeneous cell populations [67].
- Implement droplet digital PCR (ddPCR) for highly sensitive quantification of editing efficiencies, especially for HDR events.
- Utilize next-generation sequencing for comprehensive characterization of editing outcomes, particularly for detecting complex modifications or low-frequency events [9].
- Selection Strategies:
Frequently Asked Questions (FAQs)
Q1: Which genome editing technology is most suitable for large-scale screening applications? CRISPR-Cas9 is generally preferred for large-scale screening due to its simplicity in library generation and scalability. The ability to synthesize numerous guide RNAs in parallel enables genome-wide loss-of-function screens with comprehensive coverage. Additionally, the integration of CRISPR with single-cell RNA sequencing technologies enables high-resolution analysis of perturbation effects at unprecedented scale and resolution [95].
Q2: How do I choose between CRISPR, TALENs, and ZFNs for therapeutic applications? Therapeutic application requires careful consideration of multiple factors. CRISPR offers advantages in multiplexing and ease of design but may present greater off-target concerns. TALENs provide high specificity and have demonstrated success in clinical trials, particularly for ex vivo applications like CAR-T cell engineering. ZFNs have established clinical validation and smaller coding size advantageous for viral delivery, though design complexity can be limiting. The choice depends on the specific therapeutic context, target sequence constraints, and delivery considerations [94] [95] [96].
Q3: What are the key advancements in CRISPR technology that address early limitations? Recent innovations have substantially improved CRISPR capabilities:
- Base editing enables direct chemical conversion of nucleotides without double-strand breaks, expanding precision editing possibilities [95] [70].
- Prime editing offers even greater versatility, supporting all possible base substitutions, insertions, and deletions without requiring donor templates or double-strand breaks [70].
- Advanced delivery systems like LNP-SNAs improve efficiency and reduce toxicity [13].
- AI-integrated design tools enhance gRNA selection and minimize off-target effects through predictive modeling [70].
- Expanded PAM recognition through engineered Cas variants broadens the targeting range [95].
Q4: What experimental controls are essential for genome editing experiments? Proper controls are critical for interpreting editing outcomes:
- Negative controls: Cells transfected with non-targeting gRNA or inactive nucleases to account for background effects and off-target activity [9].
- Positive controls: Well-characterized gRNAs or editing systems known to function effectively in your experimental system [9].
- Untreated controls: Wild-type cells to establish baseline genetic and phenotypic profiles.
- Multiple gRNAs: When using CRISPR, include multiple guide RNAs targeting the same gene to control for guide-specific artifacts [65].
Q5: How can I improve HDR efficiency for precise gene editing? Enhancing HDR requires strategic experimental design:
- Cell cycle synchronization: HDR occurs primarily in S/G2 phases, so synchronizing cells can improve efficiency.
- Suppressing NHEJ pathways: Small molecule inhibitors (e.g., KU-0060648) can temporarily suppress competing NHEJ repair mechanisms.
- Optimized donor design: Incorporate homologous arms with appropriate length and consider single-stranded versus double-stranded donor templates.
- Cas9 engineering: Use Cas9 variants that favor HDR or time nuclease expression to coincide with donor template availability.
Research Reagent Solutions: Essential Materials
Table 2: Key reagents and resources for genome editing experiments
| Reagent Category | Specific Examples | Function & Application | Selection Considerations |
|---|---|---|---|
| Nuclease Platforms | SpCas9, FokI-dCas9, ZFN arrays | Core editing components; induce targeted DNA breaks | PAM requirements, specificity, size constraints for delivery |
| Delivery Systems | LNP-SNAs [13], viral vectors (AAV, lentivirus), electroporation | Introduce editing components into cells | Efficiency, cytotoxicity, tropism, payload size limitations |
| Design Tools | CRISPRon [70], DeepCRISPR [70], E-CRISP [98] | Predict optimal targets and minimize off-target effects | Algorithm performance, user interface, dataset training basis |
| Detection Assays | T7EI, GUIDE-seq [98], NGS, Sanger sequencing | Validate editing efficiency and specificity | Sensitivity, throughput, cost, quantitative capability |
| Control Reagents | Non-targeting gRNAs, inactive nucleases, target site plasmids | Experimental validation and benchmarking | Relevance to target system, prior validation status |
| Cell Culture Resources | HDR enhancers, transfection reagents, selection antibiotics | Support editing implementation and cell recovery | Compatibility with cell type, toxicity profile, efficiency |
Experimental Protocols: Key Methodologies
High-Efficiency CRISPR Editing Protocol
This protocol incorporates recent advancements in CRISPR technology to maximize editing efficiency while minimizing off-target effects, specifically designed for human cell line applications:
Guide RNA Design and Selection:
- Utilize AI-based prediction tools (e.g., DeepCRISPR, CRISPRon) to identify optimal guide RNAs with high predicted on-target activity and low off-target potential [70].
- Select 3-4 candidate guide RNAs targeting different regions of your gene of interest to account for variability in experimental performance.
- Verify uniqueness of the 12-nucleotide seed sequence adjacent to the PAM to minimize off-target binding.
Delivery System Preparation:
- Prepare LNP-SNAs according to published protocols [13]: Encapsulate Cas9 mRNA or protein and sgRNA within lipid nanoparticles coated with spherical nucleic acids.
- Alternatively, for hard-to-transfect cells, consider ribonucleoprotein (RNP) complex delivery by pre-incubating purified Cas9 protein with sgRNA for 15-20 minutes before transfection.
Cell Transfection and Editing:
- Transfect cells at 50-70% confluence using optimized parameters for your specific cell type.
- For LNP-SNAs, use a concentration range of 10-50 μg/mL based on preliminary optimization experiments.
- Include controls: non-targeting sgRNA for background assessment and a validated positive control sgRNA for efficiency benchmarking.
Efficiency Maximization Strategies:
- For HDR-mediated editing, include HDR enhancers such as 1-2 μM RS-1 during and after transfection.
- Implement temperature optimization (32°C post-transfection) for certain cell types to enhance editing efficiency.
- Consider Cas9 engineering: Use high-fidelity variants (e.g., eSpCas9) for reduced off-target effects or xCas9 for expanded PAM recognition when needed [95].
Validation and Analysis:
- Harvest cells 48-72 hours post-transfection for initial efficiency assessment.
- Perform T7 endonuclease I assay or Tracking of Indels by DEcomposition (TIDE) analysis for rapid efficiency quantification.
- Conduct targeted next-generation sequencing for comprehensive characterization of editing outcomes and off-target assessment.
- For clonal analysis, perform single-cell sorting and expand colonies for molecular validation.
Off-Target Assessment Methodology
Comprehensive evaluation of editing specificity is essential for rigorous genome editing applications:
In Silico Prediction:
- Utilize multiple bioinformatic tools (e.g., Cas-OFFinder, CCTop, COSMID) to predict potential off-target sites with up to 5 nucleotide mismatches [98].
- Pay particular attention to sites with mismatches in the PAM-distal region, as these are more readily tolerated by wild-type SpCas9.
Empirical Detection:
- Implement GUIDE-seq for genome-wide off-target profiling: Transfect cells with Cas9-sgRNA RNP complexes along with GUIDE-seq oligonucleotides, followed by library preparation and sequencing [98].
- For in vitro assessment, consider CIRCLE-seq: Isolate genomic DNA, incubate with Cas9-sgRNA complexes, and sequence cleavage sites with single-nucleotide resolution [98].
- Alternatively, utilize DISCOVER-Seq for in vivo applications, which leverages MRE11 binding to identify Cas9 cleavage sites [98].
Validation and Reporting:
- Amplify and sequence all predicted and empirically identified off-target sites using amplicon sequencing.
- Report off-target rates as a percentage of total reads showing indels at each site.
- Compare off-target profiles across different delivery methods and Cas9 variants to optimize specificity.
The optimal genome editing platform varies significantly based on application requirements, target constraints, and experimental context. CRISPR technology offers unparalleled simplicity, scalability, and multiplexing capability, making it ideal for high-throughput screening and applications where rapid iteration is valuable. TALENs provide exceptional specificity and remain advantageous for targets with sequence constraints that challenge CRISPR systems. ZFNs offer the smallest coding size, facilitating delivery in viral vector systems, and have established clinical validation. Emerging enhancements, particularly AI-driven design tools and advanced delivery systems like LNP-SNAs, continue to address limitations and expand the capabilities of all platforms. By understanding the comparative strengths and limitations of each system, researchers can make informed decisions to advance their genetic research and therapeutic development goals.
Conclusion
Optimizing CRISPR efficiency requires an integrated approach combining AI-driven design, sophisticated delivery systems, and cell-type specific understanding of DNA repair mechanisms. The emergence of AI-designed editors like OpenCRISPR-1 demonstrates the transformative potential of machine learning in creating highly functional tools that overcome natural evolutionary constraints. Future directions must focus on developing more predictive cellular models, advancing delivery precision to specific tissues, and establishing standardized validation frameworks that bridge laboratory findings with clinical applications. As CRISPR technologies mature toward broader therapeutic implementation, the systematic optimization of editing efficiency remains paramount for both fundamental research and successful clinical translation in genetic medicine.
References
- 1. The promises of large language for models and... protein design [https://pmc.ncbi.nlm.nih.gov/articles/PMC10701588/]
- 2. -Powered AI Unlocked Through Text-Guided Protein Editing Design [https://www.synbiobeta.com/read/ai-powered-protein-editing-unlocked-through-text-guided-design]
- 3. New Strategy Enhances CRISPR Editing Efficiency for Therapeutic Use [https://www.genengnews.com/topics/genome-editing/new-strategy-enhances-crispr-editing-efficiency-for-therapeutic-use/]
- 4. -Driven AI Engineering for Protein Therapy Advancements Gene [https://www.azolifesciences.com/news/20250423/AI-Powered-Protein-Engineering-for-Precise-and-Scalable-Gene-Therapy.aspx]
- 5. Frontiers | Strategies for CRISPR -based knock-ins in primary human... [https://www.frontiersin.org/journals/immunology/articles/10.3389/fimmu.2025.1589729/full]
- 6. - AI could lead to faster powered therapies, Stanford... CRISPR gene [https://med.stanford.edu/news/all-news/2025/09/ai-crispr-gene-therapy.html]
- 7. delivery for drug Optimizing | Drug CRISPR News discovery Discovery [https://www.drugdiscoverynews.com/optimizing-crispr-delivery-for-drug-discovery-16487]
- 8. : Revolutionizing Gene Editing & Medicine CRISPR 2025 [https://researchmatics.com/articles/crispr-2025-revolutionizing-gene-editing-medicine]
- 9. for Common Troubleshooting -Cas9 Editing Problems Guide CRISPR [https://synapse.patsnap.com/article/troubleshooting-guide-for-common-crispr-cas9-editing-problems]
- 10. Advancing CRISPR genome editing into gene therapy clinical trials: progress and future prospects - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC12094669/]
- 11. : From CRISPR To Clinic—And Charting A Path Forward Discovery [https://www.forbes.com/councils/forbestechcouncil/2025/02/13/crispr-from-discovery-to-clinic-and-charting-a-path-forward/]
- 12. CRISPR Clinical Trials: A 2025 Update - Innovative Genomics Institute (IGI) [https://innovativegenomics.org/news/crispr-clinical-trials-2025/]
- 13. Scientists just made CRISPR three times more effective | ScienceDaily [https://www.sciencedaily.com/releases/2025/09/250907024543.htm]
- 14. -Based Genome Editing Support— CRISPR Troubleshooting [https://www.thermofisher.com/de/de/home/technical-resources/technical-reference-library/genome-editing-support-center/crispr-based-genome-editing-support/crispr-based-genome-editing-support-troubleshooting.html]
- 15. Scientists just made gene editing far more powerful | ScienceDaily [https://www.sciencedaily.com/releases/2025/10/251025084545.htm]
- 16. Highly accurate protein structure prediction with AlphaFold | Nature [https://www.nature.com/articles/s41586-021-03819-2]
- 17. Frontiers | Before and after AlphaFold2: An overview of protein structure prediction [https://www.frontiersin.org/journals/bioinformatics/articles/10.3389/fbinf.2023.1120370/full]
- 18. Programmable RNA base editing with photoactivatable CRISPR -Cas13 [https://www.nature.com/articles/s41467-024-44867-2?error=cookies_not_supported&code=b1221f7d-96aa-4949-93d8-71e78728424b]
- 19. of Optimization /Cas12f1 guide RNAs using CRISPR 3 for... AlphaFold [https://colab.ws/articles/10.1016%2Fj.microc.2025.113194]
- 20. Design of highly functional genome editors by modelling CRISPR–Cas sequences | Nature [https://www.nature.com/articles/s41586-025-09298-z]
- 21. Rethinking Protein Drug Design with Highly Accurate Structure Prediction of Anti-CRISPR Proteins - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8949011/]
- 22. De novo protein design by inversion of the AlphaFold structure prediction network - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10204179/]
- 23. AI Meets CRISPR : The Future of Smart Gene - Poniak Times Editing [https://www.poniaktimes.com/ai-crispr-gene-editing-2025/]
- 24. Addgene: CRISPR Guide [https://www.addgene.org/guides/crispr/]
- 25. Unlock CRISPR Gene Editing Potential: PAM | IDT sequences [https://eu.idtdna.com/page/support-and-education/decoded-plus/crispr-cas-nucleases-understanding-pam-requirements-and-cas-diversification-strategies/]
- 26. News: OpenCRISPR-1: Generative AI Meets CRISPR - CRISPR Medicine [https://crisprmedicinenews.com/news/opencrispr-1-generative-ai-meets-crispr/]
- 27. Gene Editing CRISPR [https://www.linkedin.com/pulse/crispr-gene-editing-luke-mclaughlin-kbejf]
- 28. Frontiers | Efficiency Optimization of CRISPR/Cas9-Mediated Targeted ... [https://www.frontiersin.org/articles/10.3389/fpls.2019.00612/full]
- 29. Rapid two-step target capture ensures efficient -Cas9-guided... CRISPR [https://pubmed.ncbi.nlm.nih.gov/40273916/]
- 30. CRISPR-Based Genome Editing Support—Troubleshooting | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/technical-resources/technical-reference-library/genome-editing-support-center/crispr-based-genome-editing-support/crispr-based-genome-editing-support-troubleshooting.html]
- 31. Methods for Optimizing CRISPR-Cas9 Genome Editing Specificity - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC4976696/]
- 32. Therapeutic in vivo delivery of gene editing agents - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9454337/]
- 33. CRISPR’s breakthrough problem [https://cen.acs.org/articles/95/i7/CRISPRs-breakthrough-problem.html]
- 34. Lipid Nanoparticles vs. Viral Vectors: A Comparison for Gene Therapy [https://www.helixbiotech.com/post/lipid-nanoparticles-vs-viral-vectors-a-comparison-for-gene-therapy]
- 35. Researchers Upgrade A New Generation of Virus -Like Particles with... [https://www.creative-biogene.com/blog/researchers-upgrade-a-new-generation-of-virus-like-particles-with-higher-production-efficiency-and-delivery-efficiency]
- 36. Virus-like Particles as Nanocarriers for Intracellular Delivery of Biomolecules and Compounds - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9503347/]
- 37. Base Editing and Prime Editing | SpringerLink [https://link.springer.com/chapter/10.1007/978-3-031-46150-7_2]
- 38. : Technology and Approaches for High Optimizing ... CRISPR Efficiency [https://en.vectorbuilder.com/resources/vector-academy/gene-delivery-spotlight/optimizing-crispr.html]
- 39. Back to Basics - Base & Prime Editing - Front Line Genomics [https://frontlinegenomics.com/back-to-basics-base-and-prime-editing/]
- 40. of DNA targeting Comparison in human cells - PMC CRISPR editors [https://pmc.ncbi.nlm.nih.gov/articles/PMC9844007/]
- 41. -based functional genomics screening in... CRISPR [https://pmc.ncbi.nlm.nih.gov/articles/PMC10203043/]
- 42. Genome-wide CRISPR /Cas9 screening in human iPS derived... [https://www.nature.com/articles/s41598-021-92988-1?error=cookies_not_supported&code=66c09083-da21-4849-8bbc-c5034582a692]
- 43. CRISPR-GRANT: a cross-platform graphical analysis tool for high-throughput CRISPR-based genome editing evaluation | BMC Bioinformatics | Full Text [https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-023-05333-w]
- 44. CRISPRMatch: An Automatic Calculation and Visualization Tool for High-throughput CRISPR Genome-editing Data Analysis [https://www.ijbs.com/v14p0858.htm]
- 45. What is high - throughput (HTS) and how can... | Lonza screening [https://bioscience.lonza.com/lonza_bs/CH/en/what-is-high-throughput-screening-and-how-can-lonza-help]
- 46. - High for optimizing adoptive T cell therapies throughput screening [https://ehoonline.biomedcentral.com/articles/10.1186/s40164-024-00580-w]
- 47. /Cas9 Formats for Highly Efficient and CRISPR - High Cell... Throughput [https://www.thermofisher.com/ca/en/home/life-science/genome-editing/genome-editing-learning-center/genome-editing-resource-library/genome-editing-application-notes/crispr-cas9-formats-highly-efficient-high-throughput-cell-engineering-poster.html]
- 48. A high - throughput strategy for detecting... screening [https://pubmed.ncbi.nlm.nih.gov/25409780/]
- 49. CRISPR-Based Genome Editing Support | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/technical-resources/technical-reference-library/genome-editing-support-center/crispr-based-genome-editing-support.html]
- 50. for Optimizing -Cas9 Gene Editing sgRNA Design CRISPR [https://lifesciences.danaher.com/us/en/library/sgrna-crispr-design.html]
- 51. Cas9 Crispr | Sgrna DesignTool | Synbio... Design Sgrna [https://synbio-tech.com/sgrna-design-center]
- 52. Principles for Optimal sgRNA Design CRISPR Efficiency [https://synapse.patsnap.com/article/sgrna-design-principles-for-optimal-crispr-efficiency]
- 53. Designing Guide RNAs [https://www.takarabio.com/learning-centers/gene-function/gene-editing/gene-editing-tools-and-information/sgrna-design-tools]
- 54. -Based Genome Editing Support— CRISPR Troubleshooting [https://www.thermofisher.com/fr/fr/home/technical-resources/technical-reference-library/genome-editing-support-center/crispr-based-genome-editing-support/crispr-based-genome-editing-support-troubleshooting.html]
- 55. -GPT helps scientists to generate designs, CRISPR data and... analyze [https://www.news-medical.net/news/20250916/CRISPR-GPT-helps-scientists-to-generate-designs-analyze-data-and-troubleshoot-design-flaws.aspx]
- 56. Mastering Transfection Techniques [https://www.numberanalytics.com/blog/mastering-transfection-techniques-microbial-genetics]
- 57. DNA Transfection Troubleshooting | GenScript [https://www.genscript.com/dna-transfection-troubleshooting-guide.html]
- 58. Lipid-Based Transfection—Troubleshooting | Thermo Fisher Scientific - US [https://www.thermofisher.com/us/en/home/technical-resources/technical-reference-library/transfection-support-center/lipid-transfection-support/lipid-transfection-support-troubleshooting.html]
- 59. Cause of CRISPR Failure Identified and Reversed [https://www.genengnews.com/topics/genome-editing/cause-of-crispr-failure-identified-and-reversed/]
- 60. Biochemists discover cause of genome editing failures with hyped CRISPR system | UIC today [https://today.uic.edu/biochemists-discover-cause-of-genome-editing-failures-with-hyped-crispr-system/]
- 61. A Flow Cytometric Method to Determine Transfection Efficiency [https://bio-protocol.org/en/bpdetail?id=3244&type=0]
- 62. – Mirus Bio Transfection Efficiency [https://www.mirusbio.com/transfection-efficiency/]
- 63. Genome-scale CRISPR screening for modifiers of cellular LDL uptake [https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1009285]
- 64. - CRISPR Editing: Prediction, Analysis, and More Off Target [https://www.synthego.com/blog/crispr-off-target-editing/]
- 65. Troubleshooting CRISPR | Harvard Medical School [https://hms.harvard.edu/news/troubleshooting-crispr]
- 66. -Cas9 CRISPR - Off : Challenges and Solutions Target Effects [https://www.azolifesciences.com/article/CRISPR-Cas9-Off-Target-Effects-Challenges-and-Solutions.aspx]
- 67. CRISPR-Cas9 Troubleshooting | Blog | ZAGENO [https://blog.zageno.com/crispr-cas9-troubleshooting]
- 68. Latest Developed Strategies to Minimize the Off - Target in... Effects [https://pmc.ncbi.nlm.nih.gov/articles/PMC7407193/]
- 69. Mastering CRISPR -Cas9 for Gene Editing [https://www.numberanalytics.com/blog/mastering-crispr-cas9-gene-editing]
- 70. Revolutionizing CRISPR technology with artificial intelligence | Experimental & Molecular Medicine [https://www.nature.com/articles/s12276-025-01462-9]
- 71. Revolution of Biotechnology with CRISPR | Experimental & Molecular Medicine [https://www.nature.com/articles/s12276-025-01452-x]
- 72. Neuronal DNA reveals strategies to influence repair editing... CRISPR [https://pmc.ncbi.nlm.nih.gov/articles/PMC11230251/]
- 73. Strategies for High-Efficiency Mutation Using the CRISPR/Cas System - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC8860194/]
- 74. How to Improve CRISPR | IDT Genome Editing Efficiency [https://sg.idtdna.com/pages/support/faqs/my-crispr-genome-editing-efficiency-seems-very-low.-are-there-any-steps-i-can-take-to-improve-this]
- 75. Confidence: The Power of Controls in CRISPR Genome Editing [https://www.editco.bio/blog/crispr-confidence-the-power-of-controls-in-genome-editing]
- 76. CRISPRMatch: An Automatic Calculation and Visualization Tool for High-throughput CRISPR Genome-editing Data Analysis - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC6036748/]
- 77. CRISPR 101: Validating Your Genome Edit [https://blog.addgene.org/crispr-101-validating-your-genome-edit]
- 78. How To Use ICE: A Detailed Guide for Analyzing CRISPR Editing Results [https://www.synthego.com/guide/how-to-use-crispr/ice-analysis-guide/]
- 79. Analyzing CRISPR Editing Results with ICE from Synthego [https://blog.addgene.org/analyzing-crispr-editing-results-with-ice-from-synthego]
- 80. Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR | Genome Biology | Full Text [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-015-0843-6]
- 81. CRISPR Validation FAQ Frequently Asked Questions | GENEWIZ [https://web.genewiz.com/crispr-faq]
- 82. 5 tips when starting a CRISPR -Cas9 experiment | IDT [https://www.idtdna.com/page/support-and-education/decoded-plus/quick-tips-when-starting-a-crispr-experiment/]
- 83. Low Knockout Efficiency in Troubleshooting CRISPR Experiments [https://www.biosynsis.com/blog/troubleshooting-low-knockout-efficiency-in-crispr-experiments/]
- 84. Regulation of DNA throughout the repair cycle | Nature Reviews... cell [https://www.nature.com/articles/nrm2351?error=cookies_not_supported&code=c1d42ae2-f485-490d-a505-cc463341b165]
- 85. and mechanisms | Radiobiology... | Fiveable DNA repair pathways [https://fiveable.me/radiobiology/unit-5/dna-repair-pathways-mechanisms/study-guide/E3q2gK1WYDN4cuQH]
- 86. 5 tips when starting a CRISPR-Cas9 experiment | IDT [https://www.idtdna.com/pages/community/blog/post/quick-tips-when-starting-a-crispr-experiment]
- 87. mechanisms in DNA and repair - dividing non dividing cells [https://pubmed.ncbi.nlm.nih.gov/23684800/]
- 88. Comparative Analysis of Methods for Assessing On-Target Gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11932317/]
- 89. Overcoming Transfection Disconnection: Optimize Your CRISPR ... [https://www.synthego.com/blog/crispr-optimization]
- 90. Advancements in CRISPR Screening Techniques - Simple Science [https://scisimple.com/en/articles/2025-11-04-advancements-in-crispr-screening-techniques--a3jxnjd]
- 91. Streamlined, single-step non-viral CRISPR -Cas9 knockout strategy... [https://arthritis-research.biomedcentral.com/articles/10.1186/s13075-024-03294-w]
- 92. -based functional genomics in human dendritic cells | eLife CRISPR [https://elifesciences.org/articles/65856]
- 93. CRISPR Efficiency Prediction Documentation | DRSC/TRiP Functional Genomics Resources & DRSC-BTRR [https://fgr.hms.harvard.edu/crispr-efficiency-documentation]
- 94. Genome Editing Market Report 2025 : CRISPR and Beyond - Emerging... [https://finance.yahoo.com/news/genome-editing-market-report-2025-151500990.html]
- 95. Frontiers | Unraveling the future of genomics: CRISPR, single-cell omics, and the applications in cancer and immunology [https://www.frontiersin.org/journals/genome-editing/articles/10.3389/fgeed.2025.1565387/full]
- 96. , ZFN and TALEN /Cas-based methods for genome... CRISPR [https://pmc.ncbi.nlm.nih.gov/articles/PMC3694601/]
- 97. Gene Editing Techniques: ZFNs, TALENs or CRISPR ? [https://www.news-medical.net/life-sciences/Gene-Editing-Techniques-ZFNs-TALENs-or-CRISPR.aspx]
- 98. Benchmarking and integrating genome-wide CRISPR off-target detection and prediction - PMC [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7672467/]