CRISPR-Cas9 Genome Editing: Principles, Clinical Applications, and 2025 Outlook for Researchers
This article provides a comprehensive overview of the basic principles of CRISPR-Cas9 genome editing, tailored for researchers, scientists, and drug development professionals.
CRISPR-Cas9 Genome Editing: Principles, Clinical Applications, and 2025 Outlook for Researchers
Abstract
This article provides a comprehensive overview of the basic principles of CRISPR-Cas9 genome editing, tailored for researchers, scientists, and drug development professionals. It explores the fundamental mechanisms of RNA-guided DNA targeting, double-strand break repair pathways, and the evolution of CRISPR systems from bacterial immunity to therapeutic applications. The content covers current methodological approaches including ex vivo and in vivo editing strategies, base editing, and prime editing technologies, while addressing critical challenges such as off-target effects, delivery limitations, and immune responses. Finally, it examines the validation framework through clinical trial progress and the transformative impact of AI on CRISPR optimization, synthesizing key developments that are shaping the future of gene therapy and precision medicine.
The CRISPR-Cas9 Blueprint: From Bacterial Immunity to Programmable Genome Engineering
The discovery of the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) system and its development into the CRISPR-Cas9 gene-editing technology represents one of the most significant scientific breakthroughs of the 21st century. This revolutionary technology originated from the study of a simple bacterial immune defense mechanism and has since transformed nearly every field of biological research, from basic science to therapeutic development [1] [2]. The journey from fundamental bacteriological research to a precise genome-editing tool exemplifies how curiosity-driven science can yield transformative technologies with far-reaching implications. This whitepaper traces the historical discovery of CRISPR-Cas9, details its molecular mechanisms, classifies its system variants, and explores its applications in biomedical research and drug development, providing researchers with a comprehensive technical resource on this groundbreaking technology.
Historical Timeline of Key Discoveries
The development of CRISPR-Cas9 from an obscure bacterial sequence to a revolutionary gene-editing tool spanned nearly three decades of international scientific effort. The table below chronicles the pivotal discoveries that enabled this transformation.
Table 1: Historical Timeline of Key CRISPR-Cas Discoveries
| Year | Discovery | Key Researchers/Teams | Significance |
|---|---|---|---|
| 1987 | Identification of unusual repetitive sequences in E. coli | Yoshizumi Ishino et al. [3] | First accidental discovery of CRISPR sequences |
| 1993-2005 | Characterization of CRISPR loci and function | Francisco Mojica et al. [4] | Recognized CRISPR as a distinct family; hypothesized adaptive immune function |
| 2005 | Spacer sequences match bacteriophage DNA; PAM identification | Mojica et al.; Bolotin et al. [4] | Confirmed CRISPR as adaptive immune system; discovered PAM requirement |
| 2006 | Hypothetical scheme for CRISPR as bacterial immune system | Eugene Koonin et al. [4] | Computational prediction of CRISPR immune function |
| 2007 | Experimental demonstration of adaptive immunity | Philippe Horvath et al. [4] | Showed CRISPR integrates new phage DNA and provides resistance |
| 2008 | CRISPR spacers transcribed into guide RNAs | John van der Oost et al. [4] | Identified crRNAs that guide Cas proteins to targets |
| 2008 | CRISPR acts on DNA targets | Marraffini & Sontheimer [4] | Demonstrated DNA, not RNA, is the molecular target |
| 2010 | Cas9 cleaves target DNA | Sylvain Moineau et al. [4] | Showed Cas9 creates double-stranded breaks in target DNA |
| 2011 | Discovery of tracrRNA | Emmanuelle Charpentier et al. [4] | Identified tracrRNA essential for Cas9 system |
| 2011 | CRISPR functions heterologously in other species | Virginijus Siksnys et al. [4] | Demonstrated CRISPR works across species boundaries |
| 2012 | Biochemical characterization; single-guide RNA engineering | Siksnys et al.; Doudna & Charpentier [4] | Purified Cas9 complex; created simplified sgRNA system |
| 2013 | CRISPR adapted for genome editing in eukaryotic cells | Feng Zhang et al.; George Church et al. [4] | First demonstration of CRISPR editing in human and mouse cells |
The initial discovery phase began in 1987 when Japanese researcher Yoshizumi Ishino and colleagues accidentally cloned unusual repetitive sequences interspersed with spacer sequences while analyzing the iap gene in E. coli [3]. For years, these mysterious sequences remained biological curiosities without known function. Through the 1990s, researchers including Francisco Mojica documented similar structures across diverse bacteria and archaea, with Mojica ultimately proposing the acronym "CRISPR" in 2002 [4] [3]. The critical functional insight came in 2005 when Mojica and others recognized that spacer sequences matched viral DNA fragments, correctly hypothesizing that CRISPR constitutes an adaptive immune system in prokaryotes [4] [1].
The subsequent mechanistic elucidation phase revealed how this immune system operates. In 2008, van der Oost's team showed spacer sequences are transcribed into CRISPR RNAs (crRNAs) that guide Cas proteins [4], while Marraffini and Sontheimer demonstrated DNA targeting [4]. The pivotal Cas9 protein was identified by Bolotin in 2005 [4], with Moineau confirming its DNA cleavage function in 2010 [4]. Charpentier's discovery of tracrRNA in 2011 completed the understanding of the natural system [4]. The final technological transformation occurred when multiple groups recognized CRISPR's potential as a programmable gene-editing tool. In 2012, teams led by Siksnys and by Doudna and Charpentier independently reconstituted the CRISPR-Cas9 system in vitro, demonstrating programmable DNA cleavage [4] [1]. The field exploded in 2013 when Zhang and Church's labs simultaneously adapted CRISPR-Cas9 for efficient genome editing in eukaryotic cells [4], establishing the technology as a revolutionary tool for genetic engineering.
Molecular Mechanism and System Classification
The Natural CRISPR-Cas Immune System in Bacteria
In its natural context, the CRISPR-Cas system provides adaptive immunity in bacteria and archaea through a three-stage process that protects against viral infections and plasmid transfer [5] [2]:
Adaptation (Spacer Acquisition): When a virus first infects a bacterium, the Cas1-Cas2 complex recognizes and cleaves foreign DNA into short fragments called protospacers. These fragments are then integrated as new spacers into the CRISPR array within the host genome, creating a molecular memory of the infection [5] [2].
Expression and Maturation: During subsequent infections, the CRISPR locus is transcribed as a long precursor CRISPR RNA (pre-crRNA). This pre-crRNA is processed into short, mature crRNAs, each containing a single spacer sequence that serves as a guide to recognize matching viral DNA [5].
Interference: The mature crRNAs assemble with Cas proteins to form effector complexes. When these complexes encounter nucleic acids matching the crRNA spacer sequence, they cleave and destroy the invading genetic material, thus providing immunity [5].
A critical component in self/non-self discrimination is the protospacer adjacent motif (PAM), a short, specific DNA sequence adjacent to the target site in the viral genome. The PAM requirement ensures that the CRISPR system attacks only invading DNA while avoiding autoimmunity against the bacterial host's own CRISPR arrays [4] [5].
Classification of CRISPR-Cas Systems
CRISPR-Cas systems exhibit remarkable diversity and are classified into two main classes based on their effector complex architecture [5]:
Table 2: Classification of CRISPR-Cas Systems
| Class | Types | Signature Protein | Effector Complex | Target | tracrRNA Requirement |
|---|---|---|---|---|---|
| Class 1 | I, III, IV | Cas3 (Type I), Cas10 (Type III) | Multi-subunit complex | DNA (I, IV), DNA/RNA (III) | No |
| Class 2 | II, V, VI | Cas9 (Type II), Cas12 (Type V), Cas13 (Type VI) | Single protein | DNA (II, V), RNA (VI) | Yes (for most) |
Class 1 systems utilize multi-protein effector complexes and are found in both bacteria and archaea. Type I systems employ the Cascade complex for target recognition and Cas3 for DNA degradation. Type III systems target both RNA and DNA, while Type IV systems remain poorly characterized [5].
Class 2 systems utilize a single, large effector protein for interference and have been the primary focus for biotechnological applications due to their simplicity [5] [2]. Type II systems use Cas9, which requires both crRNA and tracrRNA for function and creates blunt-ended double-strand breaks in DNA. Type V systems employ Cas12 proteins, which process their own crRNAs and create staggered DNA breaks. Type VI systems utilize Cas13, which targets RNA rather than DNA [5].
The following diagram illustrates the molecular mechanism of the Type II CRISPR-Cas9 system:
Engineered CRISPR-Cas9 for Genome Editing
The transformation of CRISPR-Cas9 from a bacterial immune system to a versatile genome-editing tool required several key engineering advancements. Researchers simplified the natural two-RNA system (crRNA and tracrRNA) by creating a single-guide RNA (sgRNA) chimera, combining essential elements into one easily programmable molecule [4] [5]. The engineered system requires only two components: the Cas9 nuclease and the sgRNA, which can be programmed to target virtually any DNA sequence adjacent to a PAM [5].
When delivered into cells, the Cas9-sgRNA complex induces double-strand breaks (DSBs) at specific genomic locations. These breaks activate the cell's endogenous DNA repair machinery, primarily through two pathways [5] [6]:
Non-Homologous End Joining (NHEJ): An error-prone repair pathway that often results in small insertions or deletions (indels) at the break site, leading to gene knockouts.
Homology-Directed Repair (HDR): A precise repair pathway that uses a donor DNA template to introduce specific genetic modifications, such as point mutations or gene insertions.
The balance between these pathways varies by cell type, with NHEJ dominating in most mammalian cells and HDR occurring primarily in cycling cells [6].
Advanced CRISPR Tool Development and Technical Considerations
Engineered Cas9 Variants and Novel Editors
Beyond wild-type Cas9, researchers have developed numerous engineered variants with enhanced capabilities [5] [3]:
- High-fidelity Cas9 variants (e.g., SpCas9-HF1, eSpCas9-1.1) with reduced off-target effects through mutations that decrease non-specific DNA binding [3].
- Cas9 nickases that cut only one DNA strand, improving specificity when used in pairs [3].
- Dead Cas9 (dCas9) with inactivated catalytic domains, serving as a programmable DNA-binding platform for transcriptional regulation (CRISPRa/CRISPRi), epigenetic modification, or imaging [7] [3].
- Base editors that enable direct chemical conversion of one DNA base to another without creating DSBs, reducing indel formation [1].
- Prime editors that use a reverse transcriptase domain fused to Cas9 nickase to directly write new genetic information into target sites [1].
- Cas12 and Cas13 systems that target DNA and RNA respectively, expanding editing capabilities [5] [3].
Experimental Considerations and Controls
Robust experimental design is essential for successful CRISPR applications. Key considerations include:
- sgRNA Design: Careful selection of target sequences with minimal off-target potential and maximal on-target efficiency using validated algorithms.
- Delivery Methods: Choosing appropriate delivery systems (viral vectors, lipid nanoparticles, electroporation) based on cell type and application [1] [8].
- Controls: Including multiple sgRNAs per target, non-targeting sgRNA controls, and Cas9-only controls to account for non-specific effects.
- Validation: Confirming edits through Sanger sequencing, next-generation sequencing, and functional assays.
Table 3: Research Reagent Solutions for CRISPR-Cas9 Experiments
| Reagent Category | Specific Examples | Function/Application | Technical Considerations |
|---|---|---|---|
| Cas9 Expression Systems | Wild-type SpCas9, High-fidelity variants, Base editors | Creates DSBs at target sites; specific editing functions | Choose based on PAM requirements, specificity needs, and desired edit type |
| Guide RNA Systems | sgRNA expression vectors, crRNA-tracrRNA complexes | Targets Cas9 to specific genomic loci | Optimize sgRNA sequence with minimal off-target potential |
| Delivery Vehicles | AAV vectors, Lentiviral vectors, Lipid nanoparticles (LNPs), Electroporation | Introduces CRISPR components into cells | Consider payload size, cell type compatibility, and toxicity |
| Repair Templates | ssODNs, dsDNA donors with homology arms | Enables precise HDR-mediated editing | Design with sufficient homology arms; optimize concentration |
| Detection & Validation | T7E1 assay, TIDE analysis, NGS-based methods | Confirms editing efficiency and specificity | Use multiple orthogonal validation methods |
The following diagram illustrates a generalized workflow for a CRISPR-Cas9 genome editing experiment:
Therapeutic Applications and Clinical Translation
CRISPR-Cas9 has demonstrated remarkable potential in therapeutic applications, with the first FDA-approved CRISPR-based therapy, Casgevy, approved for sickle cell disease and transfusion-dependent beta thalassemia in 2024 [9]. Current clinical applications include:
Genetic Disorders
- Sickle Cell Disease and Beta Thalassemia: CRISPR-mediated disruption of the BCL11A gene to reactivate fetal hemoglobin production [9].
- Hereditary Transthyretin Amyloidosis (hATTR): Intellia Therapeutics' LNP-delivered CRISPR system targeting the TTR gene in the liver, showing ~90% reduction in disease-related protein levels in clinical trials [9].
- Hereditary Angioedema (HAE): CRISPR-based reduction of kallikrein protein, with clinical trials showing 86% reduction in target protein and significant reduction in attacks [9].
Oncology Applications
- CAR-T Cell Engineering: CRISPR-mediated editing of T cells to enhance antitumor activity and overcome resistance [9] [6].
- Tumor Model Generation: Creating precise cellular and animal models for cancer research and drug screening [7] [6].
- Oncogene Disruption: Direct targeting of driver oncogenes in tumors, such as EGFRvIII in glioblastoma [6].
Infectious Diseases
- HIV Resistance: Engineering CCR5 knockout in T cells to confer HIV resistance [2].
- Antiviral Strategies: Targeting viral genomes, including HIV reservoirs and HPV [2].
- CRISPR-Enhanced Phage Therapy: Developing CRISPR-equipped bacteriophages to treat antibiotic-resistant bacterial infections [9].
The therapeutic landscape continues to expand, with ongoing clinical trials in areas including cardiovascular diseases, neurodegenerative disorders, and rare genetic conditions [9].
Challenges and Future Perspectives
Despite rapid progress, several challenges remain in the broad application of CRISPR-Cas9 technology:
Technical Hurdles
- Off-target Effects: Unintended editing at similar genomic sequences remains a concern, though high-fidelity variants and improved sgRNA design have substantially mitigated this risk [2] [10].
- Delivery Efficiency: Achieving efficient, tissue-specific delivery in vivo continues to challenge clinical translation, with ongoing development of viral and non-viral delivery systems [1] [8].
- Editing Efficiency: HDR rates remain low in many therapeutically relevant cell types, particularly non-dividing cells [7].
- Immune Responses: Pre-existing immunity to bacterial Cas proteins may limit therapeutic efficacy in some patients [1].
Safety and Ethical Considerations
- Genomic Instability: Large deletions and chromosomal rearrangements have been reported at editing sites, requiring careful safety assessment [6].
- Germline Editing: Heritable genome modifications raise significant ethical concerns and are currently subject to moratoria in many countries [2].
- Regulatory Frameworks: Evolving regulations seek to balance innovation with appropriate oversight, particularly for genetically modified organisms and clinical applications [2].
Recent technological advances show promise in addressing these challenges. Novel anti-CRISPR proteins enable precise temporal control over editing activity, reducing off-target effects [10]. Advanced delivery systems, particularly lipid nanoparticles (LNPs), have demonstrated clinical success and enable redosing strategies [9]. The continued discovery of novel Cas proteins with diverse properties expands the targeting range and specificity of CRISPR systems [5] [3].
As the field progresses, CRISPR-Cas9 technology is poised to enable increasingly sophisticated applications in basic research, therapeutic development, and biotechnology. The ongoing refinement of editing precision, delivery efficiency, and safety profiles will further establish this revolutionary technology as an indispensable tool for biological research and medical innovation.
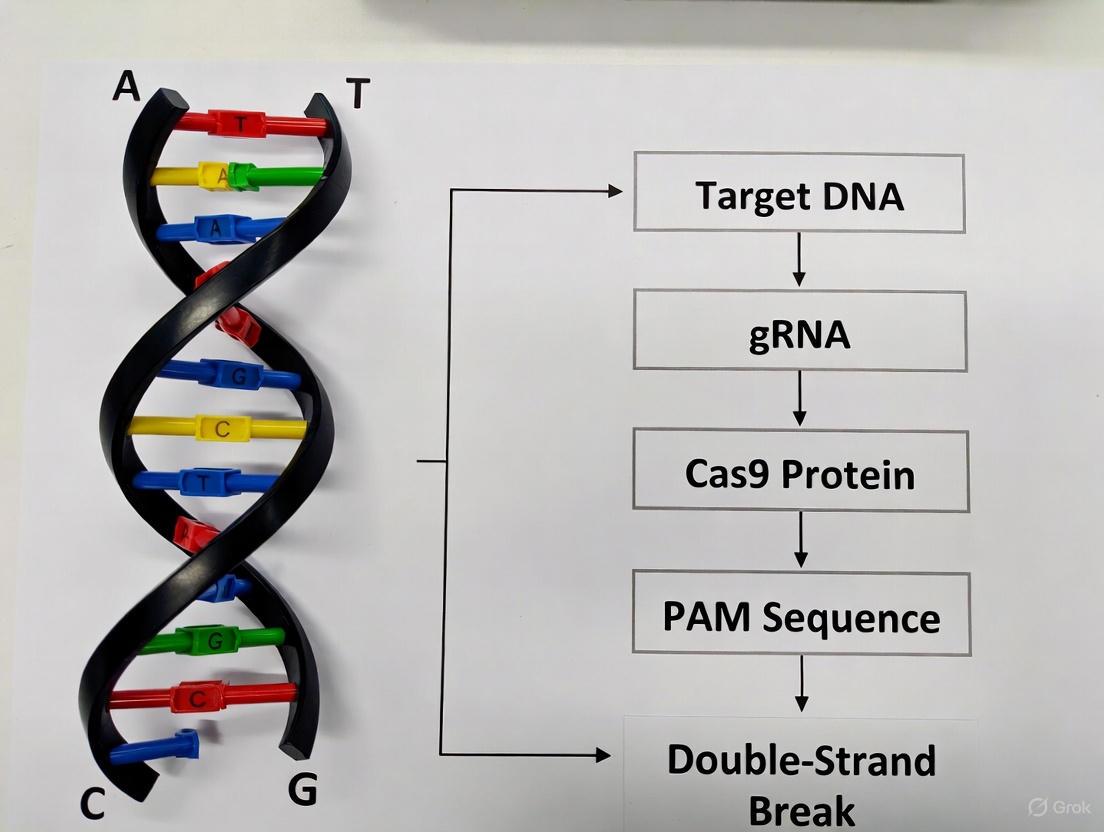
The CRISPR-Cas9 system has revolutionized genetic research and therapeutic development since its discovery, providing an unprecedented ability to perform precise genome editing [11]. This technology originates from a adaptive immune system in bacteria and archaea that protects against invading viruses and foreign genetic material [12] [13]. The core molecular machinery of this system consists of three essential components: a guide RNA (gRNA) that provides targeting specificity, a CRISPR-associated (Cas) nuclease that executes DNA cleavage, and a protospacer adjacent motif (PAM) that enables self versus non-self discrimination [14] [12]. For researchers and drug development professionals, understanding the precise structure, function, and interplay of these three components is fundamental to designing effective experiments and developing safe therapeutic applications. This technical guide examines each component in detail, providing the foundational knowledge required for advanced CRISPR-Cas9 genome editing research.
The Guide RNA (gRNA): Molecular Address System
The guide RNA serves as the targeting module of the CRISPR-Cas9 system, directing the Cas nuclease to specific genomic locations with precision. This synthetic RNA molecule combines two natural RNA components—the CRISPR RNA (crRNA) containing the target-specific spacer sequence, and the trans-activating crRNA (tracrRNA) that serves as a scaffold for Cas9 binding [15]. In engineered systems, these are often combined into a single-guide RNA (sgRNA) for simplicity [15] [16].
Structural Components and Design Principles
The gRNA contains two critical functional regions:
- Spacer Sequence: A user-defined 17-20 nucleotide sequence that determines targeting specificity through complementary base pairing with the target DNA [16]
- Scaffold Sequence: A structural component that binds to the Cas nuclease and facilitates the conformational changes required for activation [16]
The targeting specificity of the gRNA follows well-established rules. The seed sequence (8-10 bases at the 3' end of the gRNA targeting sequence) is particularly critical for initial DNA interrogation and binding [16]. Mismatches between the gRNA and target DNA in this seed region typically inhibit target cleavage, while mismatches toward the 5' end distal to the PAM are often tolerated [16].
Experimental Considerations for gRNA Design
Successful genome editing experiments require careful gRNA selection with attention to:
- Target Uniqueness: The 20-nucleotide spacer sequence must be unique compared to the rest of the genome to minimize off-target effects [16]
- Genomic Context: Accessibility of the target site within chromatin structure can significantly impact editing efficiency
- Minimizing Off-Target Effects: Bioinformatic tools are essential for predicting potential off-target sites with similar sequences [11]
Advanced applications utilize modified gRNA designs, including truncated gRNAs with shorter spacers to enhance specificity, and engineered mismatches in the PAM-distal region to promote faster Cas9 turnover after cleavage [17].
The Cas Nuclease: Molecular Scissor
The CRISPR-associated (Cas) nuclease functions as the effector module that creates double-strand breaks in target DNA. The most widely used nuclease is Cas9 from Streptococcus pyogenes (SpCas9), though numerous alternatives with distinct properties have been characterized and engineered for specialized applications [12] [16].
Structural Domains and Cleavage Mechanism
Cas nucleases exhibit a conserved bilobed architecture consisting of:
- REC Lobe (Recognition Lobe): Primarily responsible for binding the gRNA and facilitating target recognition [15]
- NUC Lobe (Nuclease Lobe): Contains the catalytic domains responsible for DNA cleavage [15]
Within the NUC lobe, two nuclease domains execute DNA cleavage:
- HNH Domain: Cleaves the target DNA strand complementary to the gRNA spacer sequence [15]
- RuvC Domain: Cleaves the non-target DNA strand [15]
These domains work coordinately to create a double-strand break approximately 3-4 nucleotides upstream of the PAM sequence [16]. The cleavage mechanism generates blunt ends for Cas9, while other nucleases like Cas12a create staggered ends with 5' overhangs [12].
Cas Nuclease Engineering and Variants
The limitations of wild-type SpCas9 have prompted extensive engineering efforts to improve its properties:
Table 1: Engineered Cas9 Variants with Enhanced Properties
| Variant Name | Key Modifications | Functional Improvements |
|---|---|---|
| eSpCas9(1.1) | Weakened interactions with non-target DNA strand | Reduced off-target effects |
| SpCas9-HF1 | Disrupted Cas9-DNA phosphate backbone interactions | Enhanced specificity |
| HypaCas9 | Increased proofreading capability | Improved mismatch discrimination |
| evoCas9 | Multiple domain mutations | Decreased off-target effects |
| xCas9 3.7 | Mutations in multiple domains | Expanded PAM recognition (NG, GAA, GAT) and increased specificity |
| Sniper-Cas9 | Not specified | Reduced off-target activity; compatible with truncated gRNAs |
| SuperFi-Cas9 | Not specified | Increased fidelity with reduced nuclease activity |
Additional Cas orthologs beyond SpCas9 offer natural alternatives with distinct properties:
Table 2: Naturally Occurring Cas Nuclease Variants
| Nuclease | Source Organism | PAM Sequence | Key Features |
|---|---|---|---|
| SaCas9 | Staphylococcus aureus | NNGRRT or NNGRRN | Smaller size for viral delivery |
| NmeCas9 | Neisseria meningitidis | NNNNGATT | Enhanced specificity |
| CjCas9 | Campylobacter jejuni | NNNNRYAC | Compact size |
| Cas12a (Cpf1) | Lachnospiraceae bacterium | TTTV | Creates staggered cuts; simpler gRNA |
| Cas12b | Alicyclobacillus acidiphilus | TTN | Thermostable variant available |
| Cas9d | Deltaproteobacteria | NGG | Compact size (747 aa); suitable for AAV delivery |
Recent structural studies of the compact Cas9d system have revealed a novel RNA-coordinated target Engagement Module (REM), where a segment of the sgRNA scaffold interacts with the REC domain to form a functional hybrid module that precisely monitors heteroduplex complementarity, resulting in lower mismatch tolerance compared to SpyCas9 [15].
The Protospacer Adjacent Motif (PAM): Self vs. Non-Self Discriminator
The protospacer adjacent motif is a short, specific DNA sequence (typically 2-6 base pairs) that follows immediately downstream of the target DNA region recognized by the gRNA [14] [12]. This sequence is absolutely required for Cas nuclease activity and serves as the primary mechanism for distinguishing between self and non-self DNA in bacterial immunity.
Biological Function and Recognition Mechanism
In native bacterial CRISPR systems, the PAM prevents autoimmunity by ensuring that Cas nucleases only target foreign DNA. While the bacterial CRISPR array contains spacer sequences derived from viruses, these sequences lack the adjacent PAM sequence, protecting the host genome from cleavage [12] [13]. The PAM recognition mechanism involves specific domains within the Cas nuclease. For SpCas9, the PAM-interacting (PI) domain recognizes the NGG motif, while in Cas9d, both the WED and PI domains collaborate in PAM recognition [15].
Structural analyses have revealed that key residues (Asn651, Lys649, and Lys715 in Cas9d) form specific hydrogen bonds with the PAM sequence, with alanine substitution of these residues abolishing or reducing target cleavage [15]. Upon PAM binding, the Cas nuclease undergoes conformational changes that destabilize the adjacent DNA duplex, enabling interrogation of sequence complementarity between the gRNA and target DNA [14].
PAM Requirements Across Cas Variants
Different Cas nucleases recognize distinct PAM sequences, which constrains their targeting ranges:
Table 3: PAM Sequences for Various CRISPR Nucleases
| CRISPR Nucleases | Organism Isolated From | PAM Sequence (5' to 3') |
|---|---|---|
| SpCas9 | Streptococcus pyogenes | NGG |
| hfCas12Max | Engineered from Cas12i | TN and/or TNN |
| SaCas9 | Staphylococcus aureus | NNGRRT or NNGRRN |
| NmeCas9 | Neisseria meningitidis | NNNNGATT |
| CjCas9 | Campylobacter jejuni | NNNNRYAC |
| StCas9 | Streptococcus thermophilus | NNAGAAW |
| LbCpf1 (Cas12a) | Lachnospiraceae bacterium | TTTV |
| AsCpf1 (Cas12a) | Acidaminococcus sp. | TTTV |
| AacCas12b | Alicyclobacillus acidiphilus | TTN |
| BhCas12b v4 | Bacillus hisashii | ATTN, TTTN and GTTN |
| Cas14 | Uncultivated archaea | T-rich PAM sequences for dsDNA cleavage |
| Cas3 | Various prokaryotic genomes | No PAM requirement |
The requirement for specific PAM sequences initially limited the targeting range of CRISPR systems, inspiring engineering efforts to develop variants with altered PAM specificities. Notable achievements include SpCas9-NG (recognizes NG PAMs), SpG (recognizes NGN PAMs), and SpRY (recognizes NRN and NYN PAMs, approaching PAM-less editing) [16].
Integrated Molecular Mechanism
The functional integration of the gRNA, Cas nuclease, and PAM recognition creates a highly specific genome editing system. The coordinated mechanism proceeds through distinct stages:
PAM Recognition and Complex Activation: The Cas nuclease scans DNA for appropriate PAM sequences, with recognition triggering conformational changes that activate the complex [14] [13]
DNA Melting and Seed Binding: PAM binding induces local DNA melting, allowing the seed region of the gRNA to interrogate potential complementarity [13] [16]
R-loop Propagation and Conformational Changes: If seed pairing is successful, the RNA-DNA heteroduplex extends, inducing structural rearrangements in the REC lobe and activating the nuclease domains [17]
Target Cleavage and Product Release: The HNH and RuvC domains create coordinated breaks in both DNA strands, after which the complex may remain bound until displaced by cellular machinery [17]
Diagram 1: CRISPR-Cas9 Target Recognition and Cleavage Mechanism
Experimental Protocols and Methodologies
In Vitro Cleavage Assay
A fundamental experiment for characterizing CRISPR-Cas9 activity involves in vitro cleavage assays:
Protocol:
- Reconstitute RNP Complex: Incubate purified Cas nuclease (1-5 μM) with synthesized gRNA (1.2-2× molar ratio) in reaction buffer (20 mM HEPES, 100 mM KCl, 5 mM MgCl₂, 1 mM DTT, pH 7.5) for 10-15 minutes at 25°C
- Add DNA Substrate: Introduce target DNA (100-500 nM) containing the appropriate PAM sequence
- Initiate Reaction: Incubate at 37°C for predetermined time points (15 minutes to several hours)
- Terminate Reaction: Add STOP buffer (10 mM EDTA, 0.1% SDS)
- Analyze Products: Separate cleavage products using agarose or polyacrylamide gel electrophoresis with appropriate DNA staining
Applications: This assay determines cleavage efficiency, kinetics, and specificity under controlled conditions, and can be adapted for high-throughput screening of gRNA efficacy or PAM specificity [18] [17].
PAM Identification Assays
Determining the PAM specificity of novel Cas nucleases requires specialized approaches:
PAM Depletion/Screening Assay:
- Library Construction: Create a plasmid library with randomized DNA sequences adjacent to a fixed target site
- Transformation: Introduce the library into bacterial cells expressing the functional CRISPR-Cas system
- Selection: Allow the system to cleave plasmids with functional PAM sequences
- Sequencing Analysis: Recover surviving plasmids and sequence the PAM region to identify depleted (functional) PAM sequences [13]
Alternative Method - PAM-SCANR: This high-throughput in vivo method uses a catalytically dead Cas variant (dCas9) coupled with a GFP reporter system. Functional PAM binding represses GFP expression, enabling FACS sorting and sequencing to identify all functional PAM motifs [13].
Research Reagent Solutions
Table 4: Essential Research Reagents for CRISPR-Cas9 Experiments
| Reagent / Tool | Function | Examples / Sources |
|---|---|---|
| Cas9 Expression Vectors | Delivery of Cas nuclease to cells | Addgene: #41815 (SpCas9), #42229 (SaCas9) |
| gRNA Cloning Vectors | Expression of single or multiple gRNAs | Addgene: #41824, #52961, #67978 |
| Cas9 Nickase Variants | Increased specificity through paired nicking | Addgene: #41816 (D10A mutant) |
| High-Fidelity Cas9s | Reduced off-target effects | eSpCas9(1.1), SpCas9-HF1, HypaCas9 |
| PAM-Flexible Variants | Expanded targeting range | xCas9, SpCas9-NG, SpRY |
| Anti-CRISPR Proteins | Inhibition of Cas9 activity after editing | LFN-Acr/PA system for rapid Cas9 inhibition |
| Bioinformatics Tools | gRNA design and off-target prediction | CHOPCHOP, CRISPResso, Cas-OFFinder |
Recent advances in reagent development include the LFN-Acr/PA system, a cell-permeable anti-CRISPR protein system that rapidly shuts down Cas9 activity after genome editing is complete, reducing off-target effects and improving clinical safety [19]. This system uses a component derived from anthrax toxin to introduce anti-CRISPR proteins into human cells within minutes, boosting genome-editing specificity by up to 40% [19].
Diagram 2: CRISPR-Cas9 Experimental Workflow
The core molecular components of the CRISPR-Cas9 system—the guide RNA, Cas nuclease, and PAM sequence—function as an integrated molecular machine that enables precise genome editing. The continuing evolution of these components through protein engineering and synthetic biology approaches is expanding the capabilities and applications of this transformative technology. For research and therapeutic development, understanding the fundamental principles governing these components and their interactions provides the foundation for designing effective experiments and developing safe genetic therapies. As the field advances, the ongoing characterization of novel Cas nucleases, refinement of gRNA design principles, and engineering of PAM specificity will further enhance the precision and utility of CRISPR-based genome editing.
The CRISPR-Cas9 system represents a paradigm shift in genome engineering, offering an unprecedented ability to perform targeted modifications within complex genomes. Its core function hinges on a fundamental biological event: the creation of a targeted double-strand break (DSB) in DNA. This controlled DNA damage is the catalyst that enables all subsequent genome editing outcomes, from gene knockouts to precise corrections. For research and drug development professionals, a deep understanding of this cutting mechanism is not merely academic; it is essential for designing effective experiments, interpreting results, and developing safe therapeutic interventions. This guide details the molecular actors, the step-by-step mechanism of DNA cleavage, and the critical experimental methodologies used to study and harness this powerful process.
Molecular Components of the CRISPR-Cas9 System
The CRISPR-Cas9 system's precision stems from its two essential components: the Cas9 nuclease and a guide RNA (gRNA). These elements work in concert to locate and cleave a specific DNA sequence.
- The Cas9 Nuclease: Cas9 is a multi-domain DNA endonuclease. Structurally, it comprises two primary lobes: the recognition (REC) lobe, which is responsible for binding the gRNA, and the nuclease (NUC) lobe [20] [21]. The NUC lobe contains two independent catalytic domains and a PAM-interaction domain:
- HNH Domain: Cleaves the DNA strand that is complementary to the gRNA (the target strand) [20] [21].
- RuvC Domain: Cleaves the non-complementary DNA strand (the non-target strand) [20] [21].
- PAM-Interacting Domain: Essential for initiating the binding to the target DNA by recognizing a short, adjacent sequence known as the Protospacer Adjacent Motif (PAM) [20] [21].
- The Guide RNA (gRNA): The gRNA is a synthetic chimeric RNA that combines the functions of two natural RNAs: the CRISPR RNA (crRNA) and the trans-activating crRNA (tracrRNA) [20] [22]. The 5' end of the gRNA contains a ~20 nucleotide spacer sequence that is complementary to the target DNA, dictating the system's specificity. The 3' end forms a hairpin structure that serves as a binding scaffold for the Cas9 protein [20] [21].
Table 1: Core Components of the CRISPR-Cas9 Cutting Machinery
| Component | Type | Key Function in DNA Cutting |
|---|---|---|
| Cas9 Nuclease | Protein (Multidomain Enzyme) | Executes the double-stranded DNA break via its HNH and RuvC nuclease domains [20] [21]. |
| Guide RNA (gRNA) | RNA Molecule | Provides sequence specificity by binding to complementary target DNA via its spacer sequence; also activates Cas9 [20] [21]. |
| Spacer Sequence | 18-20 nt region of gRNA | Determines the target genomic locus through Watson-Crick base pairing [20]. |
| Protospacer Adjacent Motif (PAM) | Short DNA sequence (e.g., 5'-NGG-3') | A mandatory recognition site adjacent to the target sequence; enables Cas9 to initiate DNA binding [20] [21] [22]. |
The Step-by-Step Mechanism of DNA Cleavage
The process of DNA cleavage by CRISPR-Cas9 is a coordinated, multi-stage mechanism that ensures high fidelity and specificity.
- Complex Assembly: The Cas9 nuclease binds to the gRNA to form a ribonucleoprotein (RNP) complex. This association induces a conformational change in Cas9, activating it for DNA surveillance [21].
- PAM Recognition and DNA Binding: The Cas9-gRNA complex scans the DNA. The PAM-interacting domain checks for the presence of a correct PAM sequence (5'-NGG-3' for the common S. pyogenes Cas9) [21] [22]. PAM recognition is a critical first step that triggers local DNA melting, unwinding the double helix adjacent to the PAM [21].
- Target Verification and R-Loop Formation: If a PAM is found, the "seed sequence" near the PAM initiates hybridization with the gRNA. If the DNA sequence is fully complementary to the gRNA spacer, the RNA-DNA hybrid extends, displacing the non-complementary DNA strand and forming a structure known as an R-loop [21].
- Conformational Activation and Double-Strand Break: Successful R-loop formation induces a final conformational change in Cas9, positioning the nuclease domains for cleavage. The HNH domain cleaves the target strand (the one hybridized to the gRNA), and the RuvC domain cleaves the non-target strand [20] [21]. This coordinated action typically results in a blunt-ended double-strand break 3 base pairs upstream of the PAM sequence [20] [21].
Diagram 1: The CRISPR-Cas9 DNA Cutting Mechanism. This workflow illustrates the sequential process from complex assembly to double-strand break formation, highlighting key steps like PAM recognition and R-loop formation.
Cellular Repair of Cas9-Induced Breaks
The DSB generated by Cas9 is not the end point of genome editing; it is the beginning of a cellular repair process. The outcome of editing is entirely determined by which of the cell's endogenous DNA repair pathways resolves the break [21] [23]. The two primary pathways are Non-Homologous End Joining (NHEJ) and Homology-Directed Repair (HDR).
Table 2: Major DNA Repair Pathways for CRISPR-Cas9-Induced Breaks
| Repair Pathway | Mechanism | Cellular Context | Typical Editing Outcome |
|---|---|---|---|
| Non-Homologous End Joining (NHEJ) | Ligates broken ends directly without a template. Error-prone [20] [21]. | Active throughout cell cycle; predominant in post-mitotic cells (e.g., neurons) [20] [23]. | Small insertions or deletions (indels); leads to gene knockouts. |
| Microhomology-Mediated End Joining (MMEJ) | Uses microhomologous sequences (5-25 bp) flanking the break for end joining. Error-prone [21]. | Active in S/G2/M phases of dividing cells [23]. | Larger deletions; distinct indel pattern. |
| Homology-Directed Repair (HDR) | Uses a homologous DNA template (donor) for precise repair [20] [21]. | Restricted to late S/G2 phases; inefficient in non-dividing cells [20] [23]. | Precise gene insertion or correction. |
Diagram 2: DNA Repair Pathway Choices After a CRISPR-Cas9 Break. The cellular machinery repairs the DSB via competing pathways, leading to different genetic outcomes, from error-prone indels to precise corrections.
Advanced Experimental Protocols for Studying DSB Mechanics
Cutting-edge research into CRISPR-Cas9 cutting and repair dynamics relies on sophisticated protocols. The following methodologies are critical for quantifying and understanding DSB mechanics in various biological contexts.
Protocol: Quantifying DSB Kinetics and Repair in Human Neurons
This protocol, adapted from a 2025 Nature Communications study, highlights the unique challenges of editing in non-dividing cells [23].
- Objective: To characterize the efficiency, outcome, and timing of CRISPR-Cas9-induced DSB repair in post-mitotic human neurons.
- Key Methodology:
- Cell Model Generation: Differentiate human induced Pluripotent Stem Cells (iPSCs) into cortical-like excitatory neurons. Validate post-mitotic state (Ki67-negative) and neuronal identity (NeuN-positive) [23].
- Cas9 Delivery via VLPs: Use Virus-Like Particles (VLPs) pseudotyped with VSVG/BRL glycoproteins to efficiently deliver pre-assembled Cas9-gRNA Ribonucleoprotein (RNP) into neurons. This method achieves high transduction efficiency without relying on cell division [23].
- Time-Course Analysis: Harvest cells at multiple time points post-transduction (e.g., days 1, 4, 7, 14). Extract genomic DNA and perform next-generation sequencing (e.g., amplicon sequencing) of the target locus.
- Data Analysis: Use computational tools (e.g., CRISPResso2) to quantify the percentage of indels and the spectrum of insertion/deletion mutations at each time point.
- Expected Results: Unlike dividing cells, where indels plateau within days, neurons show a prolonged accumulation of indels over up to two weeks, reflecting slower DSB repair kinetics and a preference for NHEJ over MMEJ pathways [23].
Protocol: Single-Molecule Dynamics of DSB Repair (UMI-DSBseq)
This method provides high-resolution, quantitative data on DSB intermediates and repair products simultaneously [24].
- Objective: To simultaneously quantify DSB intermediates and final repair products at an endogenous locus, enabling the measurement of precise repair rates.
- Key Methodology:
- RNP Delivery: Deliver pre-assembled Cas9-gRNA RNP into cells (e.g., tomato protoplasts or human cells) via PEG-mediated transformation or electroporation for synchronized DSB induction [24].
- Library Preparation (UMI-DSBseq):
- DNA Extraction: Harvest cells at multiple time points post-editing.
- End Repair: Use enzymes to create blunt ends on all DNA molecules, including unrepaired DSBs.
- Adaptor Ligation: Ligate adaptors containing Unique Molecular Identifiers (UMIs) to all blunt ends. This labels each original DNA molecule, allowing for accurate quantification and tracking of DSBs and intact molecules after sequencing [24].
- PCR Amplification & Sequencing: Amplify the target region and perform high-throughput sequencing.
- Bioinformatic Analysis: A custom computational pipeline (e.g., available on GitHub) categorizes each sequenced molecule as:
- Unrepaired DSB: Molecule with adaptor ligated at the cut site.
- Precisely Repaired: Sequence identical to the original, unedited allele.
- Error-Repaired: Contains indels [24].
- Expected Results: This technique revealed that precise repair (scar-less re-ligation) accounts for a significant majority of repair events, explaining the gap between high cleavage rates and lower observed indel frequencies [24].
The Scientist's Toolkit: Essential Reagents and Methods
Table 3: Key Research Reagents and Methods for CRISPR-Cas9 DSB Studies
| Tool / Reagent | Function/Description | Application in DSB Research |
|---|---|---|
| SpCas9 Nuclease | The standard Cas9 protein from S. pyogenes; creates blunt-ended DSBs. | The core effector protein for inducing targeted DSBs in most experimental systems [20] [21]. |
| Pre-assembled RNP | A complex of purified Cas9 protein and synthetic gRNA. | Gold standard for transient delivery; reduces off-target effects and allows for synchronized DSB induction in various cells, including primary and non-dividing cells [23] [24]. |
| Virus-Like Particles (VLPs) | Engineered particles that deliver protein cargo (e.g., Cas9 RNP) instead of genetic material. | Enables efficient delivery of CRISPR components into hard-to-transfect cells, such as neurons [23]. |
| Lipid Nanoparticles (LNPs) | Lipid-based vesicles that encapsulate and deliver CRISPR cargo. | A leading method for in vivo systemic delivery of CRISPR components, particularly to the liver [9]. |
| UMI-DSBseq | A molecular and computational toolkit using Unique Molecular Identifiers. | Enables multiplexed, single-molecule quantification of DSB intermediates and repair products over time, providing direct measurement of cutting and repair rates [24]. |
| Next-Generation Sequencing (NGS) | High-throughput DNA sequencing technologies. | The primary method for analyzing editing outcomes, including indel spectrum, efficiency, and off-target assessment via targeted amplicon sequencing. |
The creation of a targeted double-strand break is the foundational event that unlocks the full potential of CRISPR-Cas9 as a genome-editing tool. From the initial assembly of the Cas9-gRNA complex to the recognition of the PAM sequence and the final catalytic cleavage, each step is a marvel of biological precision. However, the ultimate genetic outcome is not written by CRISPR alone but is determined by the cell's own repair machinery. As research advances, the growing toolkit—from VLPs and LNPs for delivery to UMI-DSBseq for single-molecule resolution analysis—empowers scientists to dissect these mechanisms with ever-greater clarity. This deep understanding is paramount for translating CRISPR technology from a powerful laboratory technique into safe and effective therapeutic agents, paving the way for a new era in genetic medicine and drug discovery.
Non-Homologous End Joining vs. Homology-Directed Repair
In CRISPR-Cas9 genome editing, the Cas9 nuclease creates a precise double-strand break (DSB) in the DNA, but the genetic outcome is entirely determined by the cell's endogenous repair pathways [25]. The competition between two principal mechanisms—Non-Homologous End Joining (NHEJ) and Homology-Directed Repair (HDR)—represents a fundamental biological crossroads that dictates the precision and result of the edit [26]. Understanding and controlling these pathways is crucial for advancing therapeutic applications, as NHEJ is efficient but error-prone, while HDR offers precision but operates at low efficiency, particularly in non-dividing cells [23] [26].
This guide provides a technical comparison of NHEJ and HDR, details their molecular mechanisms, and synthesizes current strategies for manipulating these pathways to achieve desired editing outcomes, framing this discussion within the practical context of CRISPR-Cas9 research.
DNA Repair Pathway Fundamentals
Non-Homologous End Joining (NHEJ)
Non-Homologous End Joining (NHEJ) is an error-prone DNA repair pathway that functions throughout the cell cycle by directly ligating broken DNA ends without requiring a homologous template [25]. Its key characteristic is the frequent introduction of small insertions or deletions (indels) at the repair junction [27]. While this makes NHEJ ideal for generating gene knockouts, its lack of precision is a significant limitation for edits requiring accuracy [25]. NHEJ is the predominant and most efficient DSB repair pathway in mammalian cells [28].
Homology-Directed Repair (HDR)
Homology-Directed Repair (HDR) is a precise repair mechanism that uses a homologous DNA sequence—such as a sister chromatid or an exogenously supplied donor template—to accurately repair the DSB [25]. This pathway is essential for precise gene edits, including nucleotide substitutions, gene insertions, and the creation of tagged proteins [25] [26]. However, a major limitation is that HDR is inherently less efficient than NHEJ and is primarily active during the S and G2 phases of the cell cycle, making it particularly challenging to use in non-dividing cells [23] [26].
Table 1: Core Characteristics of NHEJ and HDR
| Feature | Non-Homologous End Joining (NHEJ) | Homology-Directed Repair (HDR) |
|---|---|---|
| Template Required | No template needed [25] | Requires homologous donor template (e.g., sister chromatid, ssDNA, dsDNA donor) [25] |
| Primary Role in CRISPR | Gene knockouts; introduction of INDELs [25] | Precise gene knock-ins; nucleotide substitutions [25] [26] |
| Fidelity | Error-prone; often results in small insertions/deletions (indels) [25] [27] | High-fidelity; enables precise, defined edits [25] |
| Efficiency | High; dominant pathway in most mammalian cells [26] [28] | Low; inefficient, especially in non-dividing cells [23] [26] |
| Cell Cycle Phase | Active throughout all cell cycle phases [23] | Primarily restricted to S and G2 phases [23] |
| Key Enzymes/ Factors | DNA-PKcs, Ku70/80, DNA Ligase IV [27] [29] | RAD51, BRCA1, BRCA2, RAD52 [28] |
Pathway Mechanics and Experimental Workflows
Visualizing the Core Pathways and Key Experiments
The following diagrams illustrate the fundamental decision between NHEJ and HDR after a CRISPR-induced break, and a key experimental workflow for studying these pathways in non-dividing cells.
Diagram 1: CRISPR Repair Pathway Decision
Diagram 2: Studying Repair in Non-Dividing Cells
Beyond NHEJ and HDR: Alternative Repair Pathways
The DSB repair landscape is more complex than the simple NHEJ-HDR dichotomy. Microhomology-Mediated End Joining (MMEJ) and Single-Strand Annealing (SSA) are two alternative, error-prone pathways that significantly contribute to imprecise editing outcomes, even when NHEJ is suppressed [28].
MMEJ relies on short microhomology sequences (2-20 base pairs) flanking the DSB for repair, typically resulting in deletions [28]. SSA requires longer homologous sequences and is mediated by Rad52, leading to deletions of the intervening sequence [28]. Studies show that inhibiting key effectors of these pathways—such as POLQ for MMEJ or Rad52 for SSA—can reduce specific imprecise integration patterns and improve the proportion of perfect HDR events [28].
Strategic Pathway Manipulation for Enhanced Editing
Quantitative Analysis of Pathway Modulation
Researchers have developed chemical and genetic strategies to shift the balance from the dominant NHEJ pathway toward HDR. The table below summarizes key small molecules used for this purpose and their quantified effects.
Table 2: Small Molecule Modulators of DNA Repair Pathways
| Small Molecule | Target/Pathway | Effect on Editing | Quantified Enhancement | Key Considerations / Risks |
|---|---|---|---|---|
| Repsox | TGF-β signaling inhibitor; promotes NHEJ [30] | Increases NHEJ-mediated knockout efficiency [30] | 3.16-fold increase in porcine cells (RNP delivery) [30] | Acts in a cell cycle-independent manner [30] |
| AZD7648 | DNA-PKcs inhibitor (NHEJ inhibitor) [27] | Intended to enhance HDR by suppressing NHEJ [27] | - | Risk: Can cause exacerbated genomic aberrations (large deletions, chromosomal translocations) [27] |
| Alt-R HDR Enhancer V2 | NHEJ pathway inhibitor [28] | Increases perfect HDR frequency in knock-in [28] | ~3-fold increase in knock-in efficiency (5.2% to 16.8%) [28] | Established method, but imprecise integration still occurs [28] |
| ART558 | POLQ inhibitor (MMEJ inhibitor) [28] | Reduces large deletions; can increase perfect HDR frequency [28] | Significant increase in perfect HDR; reduces large (≥50 nt) deletions [28] | Suppressing MMEJ can improve precision [28] |
| D-I03 | Rad52 inhibitor (SSA inhibitor) [28] | Reduces asymmetric HDR and other imprecise donor integrations [28] | Reduces specific imprecise integration patterns [28] | Effect may depend on nature of DNA cleavage ends [28] |
| Zidovudine (AZT) | Thymidine analog; suppresses HDR [30] | Enhances NHEJ-mediated gene knockout [30] | 1.17-fold increase in porcine cells [30] | - |
Optimizing Donor Template Design for HDR
The structure and delivery of the donor template are critical factors for successful HDR. Key parameters include:
- Strandedness and Orientation: Single-stranded DNA (ssDNA) donors often outperform double-stranded DNA (dsDNA). In potato protoplasts, an ssDNA donor in the "target" orientation (coinciding with the sgRNA-recognized strand) achieved the highest HDR efficiency [31].
- Homology Arm (HA) Length: While longer HAs (e.g., 300 bp to 1 kb) are traditionally used and can increase HDR efficiency [32], shorter HAs can be sufficient. Studies in potato and animal models show that ssDNA donors with HAs as short as 30-50 nucleotides can support efficient HDR or homology-mediated integration [31] [32].
- Template Delivery: Using RNP complexes (pre-assembled Cas9 protein and guide RNA) along with the donor template is a highly efficient strategy for both NHEJ and HDR, as it minimizes the time the nuclease is active and can reduce off-target effects [28] [31].
Advanced Research and Clinical Implications
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Reagents for Studying NHEJ and HDR
| Research Reagent / Tool | Function in Experimentation |
|---|---|
| Cas9 Ribonucleoprotein (RNP) | Pre-complexed Cas9 and sgRNA; enables transient editing, high efficiency, and reduced off-target effects [28] [31]. |
| Virus-Like Particles (VLPs) | Engineered particles (e.g., VSVG/BRL-pseudotyped) for efficient delivery of CRISPR components into challenging cells like human neurons [23]. |
| ssDNA Donor Template | Single-stranded oligonucleotide with homology arms; often leads to higher HDR efficiency than dsDNA, especially with short inserts [31]. |
| NHEJ Inhibitors (e.g., Alt-R HDR Enhancer V2) | Chemical suppresses the dominant NHEJ pathway to increase the relative frequency of HDR events [28]. |
| Pathway-Specific Inhibitors (ART558, D-I03) | Tools to dissect the roles of MMEJ (via POLQ inhibition) and SSA (via Rad52 inhibition) in imprecise repair outcomes [28]. |
| Long-Read Amplicon Sequencing (PacBio) | Essential for comprehensive genotyping; detects large deletions and complex structural variations missed by short-read sequencing [27] [28]. |
Addressing Genomic Stability and Safety Concerns
A pressing concern in therapeutic genome editing is the potential for on-target genomic aberrations beyond small indels. Recent studies reveal that CRISPR/Cas9 editing can lead to large structural variations (SVs), including kilobase- to megabase-scale deletions and chromosomal translocations [27]. These events are particularly aggravated by the use of certain HDR-enhancing strategies, such as DNA-PKcs inhibitors (e.g., AZD7648), which can cause a thousand-fold increase in the frequency of chromosomal translocations [27]. This underscores the critical need for advanced genotyping methods like long-read amplicon sequencing (PacBio) or assays like CAST-Seq to thoroughly assess editing outcomes and ensure patient safety [27] [28].
Cell-Type Specific Repair Responses
The choice of cell model is paramount, as DNA repair is not universal. A landmark 2025 study demonstrated that postmitotic human neurons repair Cas9-induced DSBs differently than genetically identical dividing cells (iPSCs) [23]. Neurons exhibit a much narrower distribution of indel outcomes, favor NHEJ-like repair, and accumulate edits over a prolonged period of up to two weeks, contrasting with the rapid repair seen in dividing cells [23]. This has profound implications for developing therapies for neurological diseases and highlights that editing strategies optimized in common cell lines may not translate to clinically relevant postmitotic cells.
The interplay between NHEJ and HDR forms the cornerstone of CRISPR-Cas9 genome editing outcomes. While NHEJ offers a robust tool for gene disruption, HDR holds the promise of precise genetic surgery. Current research is focused on tilting this balance toward precision by inhibiting competing pathways, optimizing donor templates, and adapting strategies to specific cell types. However, emerging challenges, such as the risk of on-target structural variations and the unique repair landscape of non-dividing cells, demand continued innovation in tool development and safety assessment. A deep understanding of these cellular repair pathways is not merely an academic exercise but a prerequisite for the safe and effective clinical translation of CRISPR-based therapies.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) proteins constitute an adaptive immune system in prokaryotes that provides sequence-specific protection against mobile genetic elements such as viruses and plasmids [33] [34]. These systems capture fragments of invading nucleic acids and incorporate them as "spacers" within CRISPR arrays in the host genome, creating a heritable genetic record of past infections [35] [34]. Upon subsequent encounters, the arrays are transcribed and processed into CRISPR RNA (crRNA) that guides Cas proteins to recognize and cleave complementary foreign nucleic acids, thereby conferring immunity [36] [33]. The natural diversity of these systems is remarkable, with CRISPR-Cas loci identified in approximately 50% of sequenced bacterial genomes and nearly 90% of sequenced archaea [34].
The classification of CRISPR-Cas systems reflects their evolutionary relationships and functional mechanisms. Systems are primarily categorized into two classes based on the architecture of their effector modules [37] [36]. Class 1 systems utilize multi-subunit effector complexes, while Class 2 systems employ single, large protein effectors [38] [39]. This fundamental distinction has profound implications for both the natural biology of these systems and their technological applications, particularly in genome editing where Class 2 systems have been more widely adopted due to their simplicity [36] [35]. Understanding this diversity provides the foundation for harnessing these systems for basic research and therapeutic development.
Classification and Genomic Organization
Hierarchical Structure of CRISPR-Cas Systems
CRISPR-Cas classification employs a multifaceted approach that combines sequence similarity, phylogenetic analysis, gene neighborhood examination, and experimental data to establish evolutionary relationships [39]. The hierarchical structure progresses from broad categories to specific variants: Class → Type → Subtype → Variant [39]. The current classification encompasses 2 classes, 7 types, and 46 subtypes, representing a significant expansion from the 6 types and 33 subtypes documented in 2020, reflecting the rapid discovery of novel systems [40].
The two-class division is based on effector complex architecture. Class 1 systems (types I, III, IV, and VII) employ multisubunit effector complexes in which different Cas proteins assemble into a complex that mediates crRNA processing and target interference [37] [40]. In contrast, Class 2 systems (types II, V, and VI) utilize a single, large multidomain effector protein for the same functions [36] [38]. This classification system continues to evolve as new variants are discovered through computational mining of genomic and metagenomic databases [40] [35].
Genomic Locus Architecture
CRISPR-Cas loci typically consist of several key components: the CRISPR array itself, composed of direct repeats alternating with variable spacers; an AT-rich leader sequence that often serves as a promoter for array transcription; and cas genes that encode the Cas proteins responsible for adaptation, expression, and interference functions [34]. The adaptation module, containing Cas1 and Cas2 proteins, is relatively conserved across most systems and is responsible for acquiring new spacers from invading DNA [36]. The effector module shows substantially greater diversity and defines the specific type and subtype of each system [36] [38].
Some CRISPR-Cas systems exist in "non-autonomous" forms that lack essential components, particularly the adaptation module genes cas1 and cas2 [36]. These systems may depend on adaptation modules encoded elsewhere in the genome or may have specialized functions that do not require spacer acquisition [36] [39]. Type IV systems represent a prominent example of this non-autonomous architecture, typically lacking adaptation modules and often being encoded on plasmids rather than chromosomal DNA [39].
Class 1 CRISPR-Cas Systems: Multi-Subunit Effector Complexes
Class 1 CRISPR-Cas systems represent the evolutionarily ancestral form of CRISPR immunity and are the most abundant in prokaryotes [39]. These systems comprise approximately 90% of all identified CRISPR-Cas loci in bacteria and archaea, with near-universal presence (close to 100%) in archaeal genomes [37] [36]. Despite their natural abundance, Class 1 systems have been less widely adopted for biotechnological applications compared to Class 2 systems, primarily due to the practical challenges of reconstituting multi-protein complexes in heterologous systems [39].
The defining feature of Class 1 systems is their utilization of multi-subunit effector complexes, often referred to as Cascade (CRISPR-associated complex for antiviral defense) complexes [34] [39]. These complexes typically consist of multiple Cas protein subunits that assemble in uneven stoichiometry to form a functional unit capable of crRNA binding, target recognition, and in some cases, nucleic acid cleavage [36]. Recent advances in genetic engineering have begun to overcome the technical challenges of working with Class 1 systems, leading to increased interest in their unique properties for specialized applications [39].
Type I Systems
Type I systems represent the most prevalent CRISPR-Cas type in nature and utilize a characteristic effector complex that recruits the Cas3 protein for DNA degradation [37] [39]. The Cascade complex varies in composition across subtypes but typically includes Cas5, Cas6, Cas7, and Cas8 proteins in various combinations [39]. Cas6 often functions as the pre-crRNA processing enzyme, cleaving the long primary transcript into individual crRNA units within the repeats [34].
A defining feature of type I systems is the Cas3 protein, which contains both helicase and nuclease activities [37] [39]. After the Cascade complex identifies and binds to a target DNA sequence complementary to the crRNA guide, it recruits Cas3, which processively degrades extended regions of DNA [39]. This mechanism results in large-scale DNA deletions rather than precise double-strand breaks, making type I systems particularly useful for applications requiring extensive genomic rearrangements [39]. Type I systems are further divided into seven subtypes (I-A through I-G) based on the specific composition of their Cascade complexes and accessory proteins [37].
Type III Systems
Type III CRISPR-Cas systems are considered among the most complex and are hypothesized to represent the evolutionary ancestor of all other CRISPR systems [39]. These systems are characterized by the presence of Cas10, a multidomain protein that serves as the large subunit of the effector complex [37] [40]. Cas10 typically contains a polymerase/cyclase domain that synthesizes cyclic oligoadenylate (cOA) second messengers upon target recognition [40].
Unlike other CRISPR types, type III systems can target both RNA and DNA, though DNA cleavage is considered their primary immune function [39]. These systems can cleave RNA directly through the effector complex or indirectly by activating non-specific RNases through the cOA signaling pathway [40]. Recent analyses have identified new subtypes (III-G, III-H, and III-I) that exhibit reductive evolution, with some losing the cOA signaling pathway or specific nuclease activities [40]. The type III systems demonstrate exceptional complexity in their regulatory mechanisms and interference capabilities.
Type IV Systems
Type IV CRISPR-Cas systems represent atypical, non-autonomous systems that lack key components of canonical CRISPR-Cas immunity [39]. These systems are typically missing adaptation modules (cas1 and cas2) and often lack functional nuclease effectors, particularly in subtypes IV-A and IV-B [39]. Type IV systems are frequently encoded on plasmids rather than bacterial chromosomes and contain a distinct Cas7-type protein as their defining feature [39].
The precise biological function of type IV systems remains enigmatic, though evidence suggests they may participate in plasmid competition or regulate conjugation by targeting specific DNA sequences [39]. Unlike most other CRISPR types, type IV systems appear to have diverged from adaptive immunity functions and may represent specialized systems for nucleic acid targeting in the context of mobile genetic element competition [39]. The IV-C subtype uniquely contains a helicase domain resembling cas10, suggesting functional diversity within this type [39].
Type VII Systems
Type VII represents the most recently classified CRISPR-Cas system, identified through deep terascale clustering of microbial genomic data [40] [39]. These systems are characterized by the presence of Cas14 (also referred to as Cas7-11 in type III-E systems), an effector protein containing a metallo-β-lactamase (β-CASP) domain [40]. Type VII loci typically lack adaptation modules and are often found associated with substituted repeats in their CRISPR arrays, suggesting infrequent spacer acquisition [40].
Structural analysis reveals that type VII effector complexes can comprise up to 12 subunits, making them among the largest Class 1 complexes [40]. Despite their classification as Class 1 systems, type VII effector complexes target RNA in a crRNA-dependent manner, with cleavage mediated by the nuclease activity of Cas14 [40]. Phylogenetic and structural evidence suggests that type VII systems evolved from type III systems through a reductive evolutionary process, retaining the RNA-targeting capability while simplifying certain complex features [40].
Class 2 CRISPR-Cas Systems: Single-Effector Proteins
Class 2 CRISPR-Cas systems are defined by their utilization of a single, large multidomain effector protein for crRNA processing and target interference [36] [38]. These systems represent approximately 10% of all identified CRISPR-Cas loci and are found almost exclusively in bacterial genomes, with no documented occurrences in hyperthermophiles [36]. The relative simplicity of Class 2 systems, particularly the requirement for only a single effector protein, has made them ideal for adaptation as genome engineering tools [36] [35].
The discovery and characterization of Class 2 effectors has expanded dramatically through computational mining of genomic and metagenomic datasets [36] [35]. By using Cas1 or CRISPR arrays as "bait" in large-scale searches, researchers have identified numerous novel Class 2 variants with diverse properties [36] [38]. This exploration has revealed that Class 2 systems have evolved on multiple independent occasions through recombination events between Class 1 adaptation modules and effector proteins acquired from distinct mobile genetic elements [36] [38].
Type II Systems
Type II systems are the best-characterized Class 2 systems, largely due to the revolutionary applications of their effector protein, Cas9, in genome editing [33] [34]. These systems typically include cas1, cas2, and cas9 genes, along with the additional RNA components tracrRNA and frequently csn2 [38]. Cas9 is a large multidomain protein that contains two distinct nuclease domains: an HNH domain that cleaves the target DNA strand complementary to the crRNA guide, and a RuvC-like domain that cleaves the non-target strand [33] [34].
A defining feature of type II systems is their requirement for two RNA components: the crRNA, which contains the guide sequence for target recognition, and the trans-activating crRNA (tracrRNA), which facilitates pre-crRNA processing and Cas9 activation [34]. In laboratory applications, these are often fused into a single-guide RNA (sgRNA) to simplify implementation [34]. Cas9 requires a specific protospacer adjacent motif (PAM) sequence adjacent to the target site, typically 5'-NGG-3' for the most commonly used Streptococcus pyogenes Cas9 [33]. Type II systems are divided into three subtypes (II-A, II-B, and II-C) based on variations in their accessory proteins and genetic architecture [39].
Type V Systems
Type V CRISPR-Cas systems utilize Cas12 (formerly known as Cpf1) as their effector protein and exhibit several distinctive features compared to Cas9 [34] [39]. Cas12 proteins contain a single RuvC-like nuclease domain that cleaves both strands of target DNA, creating staggered ends with 5' overhangs rather than the blunt ends produced by Cas9 [34]. Type V systems typically recognize T-rich PAM sequences (5'-TTTV-3') and do not require a tracrRNA for function, relying solely on crRNA for guidance [34].
The type V category has expanded to include numerous subtypes (A-I and U) with diverse properties [39]. Among these, several notable variants have been characterized: Cas12a (Cpf1) processes its own pre-crRNA arrays, enabling multiplexed targeting from a single transcript [39]; Cas12f (Cas14) represents an exceptionally small effector (400-700 amino acids) that targets single-stranded DNA [39]; and certain type V variants have been engineered as CRISPR-associated transposases (CASTs) capable of inserting large DNA fragments without creating double-strand breaks [39]. This functional diversity makes type V systems particularly valuable for specialized applications beyond standard gene editing.
Type VI Systems
Type VI systems are defined by their use of Cas13 effectors, which represent the only CRISPR systems that exclusively target RNA rather than DNA [34] [39]. Cas13 proteins contain two higher eukaryotes and prokaryotes nucleotide-binding (HEPN) domains that mediate RNA cleavage activity [38] [34]. Upon recognition of a target RNA sequence complementary to the crRNA guide, Cas13 exhibits collateral RNase activity, non-specifically cleaving nearby RNA molecules in addition to the target [34].
This collateral cleavage effect has been harnessed for diagnostic applications, most notably in the SHERLOCK (Specific High-sensitivity Enzymatic Reporter unLOCKing) platform for detecting specific nucleic acid sequences [37] [34]. Type VI systems include four subtypes (VI-A through VI-D) with variations in their targeting requirements and cleavage specificities [33]. The RNA-targeting capability of type VI systems provides a powerful approach for transcript knockdown, RNA editing, and nucleic acid detection without permanent genomic alteration [39].
Comparative Analysis of CRISPR-Cas Systems
Structural and Functional Comparison
The fundamental architectural differences between Class 1 and Class 2 CRISPR-Cas systems translate to distinct functional characteristics and biological implications. Class 1 systems, with their multi-subunit effectors, generally exhibit more complex regulation and potentially more sophisticated target recognition mechanisms [36]. The Cascade complexes of Class 1 systems often undergo conformational changes upon target binding that activate nuclease functions or recruit additional effector proteins [36]. In contrast, Class 2 systems employ a single protein that integrates all functions required for interference, resulting in simpler but potentially less regulatable activity [38].
The evolutionary distribution of the two classes reveals intriguing patterns. Class 1 systems dominate in archaea and are more prevalent in bacteria, particularly in thermophilic environments [36]. This distribution suggests possible advantages of multi-subunit effectors in certain environmental conditions or cellular contexts. Class 2 systems show a more restricted phylogenetic range, being largely confined to bacteria and absent from hyperthermophiles, indicating possible evolutionary constraints on their origin or maintenance [36].
Table 1: Comparison of Major CRISPR-Cas Types and Their Characteristics
| Class | Type | Effector Complex/Protein | Target | Signature Proteins | Key Features |
|---|---|---|---|---|---|
| Class 1 | I | Multi-subunit Cascade | dsDNA | Cas3 | Recruits Cas3 for processive DNA degradation; most common type |
| Class 1 | III | Multi-subunit complex | ssRNA/DNA | Cas10 | Targets both RNA and DNA; produces signaling molecules |
| Class 1 | IV | Multi-subunit complex | dsDNA | Distinct Cas7 | Non-autonomous; often plasmid-encoded; function not fully characterized |
| Class 1 | VII | Multi-subunit complex | RNA | Cas14 | β-CASP nuclease; evolved from type III; targets RNA |
| Class 2 | II | Cas9 | dsDNA | Cas9 | Requires tracrRNA; creates blunt-end DSBs; most widely used in editing |
| Class 2 | V | Cas12 | dsDNA | Cas12 | Creates staggered DSBs; self-processes pre-crRNA; no tracrRNA needed |
| Class 2 | VI | Cas13 | RNA | Cas13 | RNA-guided RNA cleavage; exhibits collateral activity |
Quantitative Distribution in Prokaryotes
The relative abundance of different CRISPR-Cas types reveals striking patterns in their natural distribution. Class 1 systems collectively account for approximately 90% of all identified CRISPR-Cas loci across prokaryotes, with type I systems alone representing the majority of these [37] [36]. Among Class 2 systems, type II is the most prevalent, followed by types V and VI [36]. The recently identified type VII systems appear to be relatively rare compared to the more established types [40].
Analysis of CRISPR-Cas system abundance shows a characteristic "long-tail" distribution, with the well-characterized systems being relatively common while newly discovered variants are typically rare [40]. This pattern suggests that numerous additional rare variants remain to be discovered in undersampled taxonomic groups and environments [40]. The differential distribution of CRISPR types across phylogenetic lineages and habitats reflects both evolutionary history and functional adaptation to specific ecological niches and defensive requirements.
Table 2: Natural Abundance and Distribution of CRISPR-Cas Systems
| System | Approximate Abundance | Primary Phylogenetic Distribution | Notable Subtypes/Variants |
|---|---|---|---|
| Class 1 Type I | ~60% of all systems | Bacteria and Archaea | I-A to I-G (I-E most studied) |
| Class 1 Type III | ~30% of all systems | Primarily Archaea | III-A to III-I (III-A, III-B most common) |
| Class 1 Type IV | Rare | Bacteria (plasmid-encoded) | IV-A, IV-B, IV-C |
| Class 1 Type VII | Rare | Archaea | Cas14-containing systems |
| Class 2 Type II | ~7% of all systems | Bacteria only | II-A, II-B, II-C (II-A includes SpCas9) |
| Class 2 Type V | ~2% of all systems | Bacteria only | V-A to V-I, V-U (V-A includes Cas12a/Cpf1) |
| Class 2 Type VI | ~1% of all systems | Bacteria only | VI-A to VI-D (VI-B includes Cas13b) |
Experimental Methodologies for CRISPR-Cas System Characterization
Computational Discovery Pipelines
The discovery of novel CRISPR-Cas systems has been revolutionized by computational pipelines that mine the vast amount of sequence data available in genomic and metagenomic databases [36] [35]. These pipelines typically employ a multi-step process beginning with the identification of "seed" sequences that indicate the potential presence of a CRISPR-Cas system [36]. The most common seeds are the Cas1 protein, which is highly conserved and present in most systems, or CRISPR arrays themselves, which can identify non-autonomous systems lacking adaptation modules [36].
Following seed identification, the genomic neighborhood surrounding the seed is analyzed for the presence of additional cas genes and CRISPR arrays [36] [38]. Putative effector proteins are identified based on size (>500 amino acids for Class 2 effectors) and the presence of characteristic domains such as RuvC, HNH, or HEPN [38]. Candidate loci are then compared against profiles of known systems to classify them into established types or identify novel variants [36]. This approach has led to the discovery of numerous novel systems, including additional subtypes of types V and VI, and the recently identified type VII systems [36] [40].
Figure 1: Computational Pipeline for CRISPR-Cas System Discovery. This workflow illustrates the bioinformatics approach used to identify novel CRISPR-Cas systems from genomic and metagenomic sequence data.
Functional Characterization of Novel Systems
Once a novel CRISPR-Cas system has been identified computationally, experimental characterization is essential to validate its function and determine its molecular mechanisms. The initial functional assessment typically involves heterologous expression in a model system such as E. coli to test for interference activity against target sequences [38]. This approach determines whether the system can protect against phage infection or plasmid transformation in a sequence-specific manner [38].
Detailed biochemical characterization includes in vitro reconstruction of the interference reaction using purified components to assess target recognition requirements, cleavage patterns, and cofactor dependencies [38]. For Class 2 systems, this involves expression and purification of the effector protein and synthesis of the corresponding crRNA [38]. For Class 1 systems, the process is more complex, requiring co-expression and purification of multiple subunits that assemble into the functional effector complex [39]. Molecular characterization determines key properties such as PAM requirements (for DNA-targeting systems), target specificity, cleavage kinetics, and potential collateral activity [38].
Molecular Analysis Techniques
Specific experimental approaches have been developed to characterize key aspects of CRISPR-Cas system function. PAM identification typically employs plasmid transformation assays where libraries of potential target sequences containing randomized PAM regions are tested for interference [38]. Alternatively, in vitro selection methods like SELEX can identify preferred PAM sequences by assessing protein binding to randomized DNA libraries [38].
Target cleavage specificity is often evaluated using in vitro cleavage assays with synthetic target sequences, followed by gel electrophoresis to visualize cleavage products and determine cut sites [38]. For systems with putative RNase activity, RNA cleavage assays with fluorescently labeled substrates can detect cleavage products with high sensitivity [38]. High-throughput methods like RNA-seq can comprehensively map cleavage specificity and identify potential off-target effects [38].
Table 3: Essential Research Reagents for CRISPR-Cas System Characterization
| Reagent Category | Specific Examples | Function/Application |
|---|---|---|
| Expression Vectors | pET, pBAD, mammalian expression vectors | Heterologous expression of Cas effectors and complex components |
| Guide RNA Templates | Synthetic DNA oligos, gRNA expression vectors | Provision of crRNA and tracrRNA components for target recognition |
| Target Substrates | Plasmid libraries, synthetic oligonucleotides, phage DNA | Testing interference activity and sequence requirements |
| Cell-Based Assay Systems | E. coli BL21, HEK293T, phage susceptibility assays | Functional testing in cellular environments |
| Purification Systems | Affinity tags (His-tag, GST-tag), chromatography resins | Isolation of recombinant Cas proteins and complexes |
| Detection Reagents | Fluorescent reporters, antibodies, nucleotide analogs | Monitoring cleavage activity, protein expression, and localization |
Applications in Research and Therapeutic Development
Genome Editing and Regulation
The application of CRISPR-Cas systems as programmable gene editing tools has revolutionized biological research and therapeutic development [33]. The Cas9 system from type II has been most widely adopted for these applications due to its simplicity and well-characterized mechanism [33] [34]. Standard genome editing using Cas9 involves the introduction of a double-strand break at a specific genomic location guided by a custom-designed sgRNA, followed by repair through either non-homologous end joining (NHEJ) or homology-directed repair (HDR) [33]. NHEJ typically results in gene disruption through small insertions or deletions, while HDR enables precise genetic modifications using a donor DNA template [33].
Beyond standard editing, engineered variants of Cas9 have expanded the applications of CRISPR technology. Catalytically "dead" Cas9 (dCas9) retains DNA binding capability without cleavage activity and can be fused to various effector domains to enable transcriptional regulation, epigenetic modification, or genomic imaging [33]. Base editing systems combine dCas9 with nucleotide deaminases to enable direct conversion of one DNA base to another without double-strand break formation [33]. Prime editing represents a further refinement that uses a reverse transcriptase domain fused to Cas9 to directly write new genetic information into a target site [33].
Diagnostic and Biotechnology Applications
The unique properties of various CRISPR-Cas systems have enabled diverse applications beyond genome editing. The collateral RNA cleavage activity of Cas13 (type VI) has been harnessed for highly sensitive diagnostic applications through the SHERLOCK and DETECTR platforms [37] [34]. These systems can detect attomolar concentrations of specific DNA or RNA sequences, enabling rapid, low-cost detection of pathogens, genetic mutations, or other nucleic acid biomarkers [37].
The programmable RNA targeting capability of Cas13 has also been applied to transcript engineering and knockdown in eukaryotic cells, providing an alternative to RNA interference with potentially improved specificity and efficiency [39]. Similarly, the single-stranded DNA targeting activity of Cas12f (Cas14) systems has been exploited for nucleic acid detection and potentially for anti-viral applications [39]. The diversity of naturally occurring CRISPR systems continues to provide new molecular tools with specialized properties for biotechnology applications.
Therapeutic Development and Clinical Applications
CRISPR-Cas systems have rapidly advanced into clinical applications, with the first CRISPR-based therapy, Casgevy (exagamglogene autotemcel), receiving regulatory approval for the treatment of sickle cell disease and transfusion-dependent beta thalassemia [9]. This therapy uses ex vivo CRISPR-Cas9 editing of hematopoietic stem cells to reactivate fetal hemoglobin production [9]. The success of this approach has demonstrated the therapeutic potential of CRISPR technology and paved the way for numerous additional clinical developments.
Recent advances have expanded CRISPR therapeutics to in vivo applications, particularly for liver-associated diseases [9]. Lipid nanoparticle (LNP) delivery of CRISPR components has enabled efficient targeting of hepatocytes, with clinical trials underway for hereditary transthyretin amyloidosis (hATTR), hereditary angioedema (HAE), and hypercholesterolemia [9]. Intellia Therapeutics' phase I trial for hATTR demonstrated durable reduction (>90%) of disease-associated protein levels after a single LNP-based treatment, establishing proof-of-concept for in vivo CRISPR therapy [9]. The flexibility of CRISPR systems continues to inspire novel therapeutic approaches across a widening spectrum of genetic diseases.
Figure 2: Applications of CRISPR-Cas Systems in Therapeutics and Diagnostics. The diverse natural properties of different CRISPR systems have enabled multiple therapeutic approaches and diagnostic applications.
The natural diversity of CRISPR-Cas systems represents a remarkable example of molecular evolution and provides a rich resource for biotechnological innovation. The fundamental division between Class 1 multi-subunit effectors and Class 2 single-effector proteins reflects different evolutionary solutions to the challenge of adaptive immunity in prokaryotes [37] [36]. While Class 2 systems have dominated initial applications due to their simplicity, Class 1 systems offer untapped potential for specialized functions including large DNA deletions, RNA targeting, and potentially novel mechanisms not yet discovered [39].
The continuing discovery of novel CRISPR-Cas variants suggests that the current classification represents only a fraction of the natural diversity [40]. The "long tail" of rare systems in microbial genomes and metagenomes likely contains numerous additional variants with unique properties [40]. Computational approaches will continue to drive this discovery process, while structural biology and biochemical characterization will elucidate the mechanisms of newly identified systems [36] [40]. The ongoing exploration of CRISPR diversity promises to yield new molecular tools with enhanced capabilities for research and therapeutic applications.
The rapid translation of CRISPR technology from basic discovery to clinical application demonstrates the profound impact of understanding natural system diversity [9]. As the CRISPR toolkit expands to include systems with various sizes, specificities, and functions, researchers will be able to select the optimal system for each particular application [35] [39]. This expanding capability, combined with advances in delivery and specificity, positions CRISPR technology to make increasingly significant contributions to medicine, biotechnology, and basic research in the coming years. The continued study of natural CRISPR diversity will undoubtedly yield additional surprises and opportunities for innovation.
The discovery of the CRISPR-Cas9 system represents one of the most significant breakthroughs in modern biology, earning Emmanuelle Charpentier and Jennifer Doudna the 2020 Nobel Prize in Chemistry. This in-depth technical guide examines the foundational research that transformed a bacterial immune mechanism into a programmable genome-editing tool. We detail the key experiments that elucidated the molecular mechanisms of CRISPR-Cas9, from the initial identification of CRISPR sequences in prokaryotes to the structural characterization of Cas9 and its guide RNA components. The article provides comprehensive methodological protocols for recreating essential experiments, along with quantitative comparisons of CRISPR system components. Designed for researchers, scientists, and drug development professionals, this review frames these discoveries within the broader context of CRISPR-Cas9 basic principles and their transformative impact on genetic engineering.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) systems evolved in bacteria and archaea as adaptive immune defenses against mobile genetic elements, including bacteriophages and plasmids [41] [34]. This system allows prokaryotes to acquire resistance to invading viruses by incorporating fragments of viral DNA into their CRISPR loci, which then provide guidance for subsequent targeting and cleavage of matching sequences [1]. The transformation of this biological curiosity into a revolutionary genome-editing technology required key insights into its molecular mechanisms and programmable potential.
The critical breakthrough came in 2012 when Emmanuelle Charpentier and Jennifer Doudna demonstrated that the type II CRISPR system from Streptococcus pyogenes could be reconstituted in vitro and programmed to cleave any DNA sequence of choice [42] [43]. Their work simplified the natural system by combining two RNA components - crRNA and tracrRNA - into a single-guide RNA (sgRNA), creating a two-component system consisting of Cas9 protein and sgRNA that could be easily programmed to target specific DNA sequences [41] [33]. This programmable nature, coupled with its simplicity and precision, distinguished CRISPR-Cas9 from previous genome-editing technologies like zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), which required complex protein engineering for each new target [44] [33].
Historical Context and Key Discoveries
The path to understanding CRISPR began with incidental observations that evolved into a systematic exploration of bacterial adaptive immunity. The timeline below summarizes the key discoveries that led to the development of programmable genome editing:
Initial Characterization of CRISPR Loci
The first recognition of CRISPR sequences occurred in 1987 when Japanese researchers studying the iap gene in Escherichia coli accidentally cloned an unusual set of tandem repeats with interspacing sequences [34] [1]. However, the biological significance of these sequences remained mysterious for over a decade. In 2002, the term CRISPR was formally introduced to describe these Clustered Regularly Interspaced Short Palindromic Repeats, and the associated cas genes were identified [34] [1]. A critical conceptual advance came in 2005 when three independent research groups recognized that the spacer sequences between repeats were derived from phage and plasmid DNA, suggesting a role in adaptive immunity [34] [1].
Elucidation of the Immune Mechanism
Experimental demonstration of CRISPR-Cas function as an adaptive immune system came in 2007 when Barrangou et al. showed that Streptococcus thermophilus could acquire new spacers from infecting bacteriophages and thereby gain resistance to subsequent infection [34] [1]. This established CRISPR-Cas as a heritable immune system that could be programmed through spacer acquisition. Further mechanistic insights followed, including the discovery that CRISPR arrays are transcribed and processed into short CRISPR RNA (crRNA) molecules that guide Cas proteins to complementary invading DNA [41] [1].
The tracrRNA Discovery
A pivotal moment came in 2011 when Emmanuelle Charpentier, while studying Streptococcus pyogenes, discovered a previously unknown small RNA called trans-activating crRNA (tracrRNA) [42] [43]. Her work demonstrated that tracrRNA forms a duplex with crRNA and is essential for Cas9-mediated DNA cleavage in the type II CRISPR system [41]. This discovery revealed the natural two-RNA structure that guides Cas9 to its target, setting the stage for the engineering of a simplified system.
Molecular Mechanism of the CRISPR-Cas9 System
Core Components and Their Functions
The programmable CRISPR-Cas9 system consists of two fundamental components: the Cas9 endonuclease and a single-guide RNA (sgRNA). The natural system involves three components: Cas9 protein, crRNA, and tracrRNA, but the key innovation was fusing the latter two into a single chimeric guide RNA [41] [33].
Table 1: Core Components of the CRISPR-Cas9 System
| Component | Type | Function | Key Features |
|---|---|---|---|
| Cas9 Protein | Endonuclease | DNA cleavage | Contains RuvC and HNH nuclease domains; requires PAM recognition |
| crRNA | RNA | Target recognition | Contains 20-nt spacer complementary to target DNA |
| tracrRNA | RNA | Cas9 activation | Forms duplex with crRNA; required for processing and maturation |
| sgRNA | Chimeric RNA | Combined guide | Fused crRNA+tracrRNA; simplifies system to two components |
Structural Insights into Cas9 Function
Structural studies of Streptococcus pyogenes Cas9 (SpCas9) have revealed a bilobed architecture consisting of target recognition (REC) and nuclease (NUC) domains [41]. The REC lobe is responsible for sgRNA binding and target recognition, while the NUC lobe contains the DNA cleavage activity. Within the NUC lobe, two nuclease domains perform strand-specific cutting: the HNH domain cleaves the DNA strand complementary to the crRNA spacer, while the RuvC-like domain cleaves the non-complementary strand [41] [33].
The Cas9-sgRNA complex scans DNA for protospacer adjacent motifs (PAMs), short conserved sequences adjacent to the target site. For SpCas9, the PAM sequence is 5'-NGG-3', where N is any nucleotide [41] [44]. PAM recognition triggers DNA unwinding, allowing the sgRNA spacer to form an RNA-DNA heteroduplex with its complementary target sequence. Successful pairing activates Cas9's nuclease domains, resulting in a blunt-ended double-strand break (DSB) approximately 3-4 nucleotides upstream of the PAM site [44].
DNA Repair Pathways and Editing Outcomes
The cellular response to CRISPR-Cas9-induced DNA breaks determines the final editing outcome. Two major DNA repair pathways are engaged:
Non-Homologous End Joining (NHEJ) is an error-prone repair pathway that directly ligates broken DNA ends, often resulting in small insertions or deletions (indels) at the cleavage site [41] [44]. When these indels occur within a protein-coding sequence, they can produce frameshift mutations that disrupt gene function, making NHEJ particularly useful for gene knockout applications.
Homology-Directed Repair (HDR) uses a DNA template with homology to the sequences flanking the break to enable precise genome modifications [44]. While HDR occurs at lower frequency than NHEJ in most cell types, it allows for precise gene corrections, insertions, or replacements when combined with an exogenous donor DNA template.
The Seminal 2012 Experiment: Methodology and Protocols
The groundbreaking experiment published in June 2012 by Jinek et al. demonstrated the programmable DNA cleavage capability of CRISPR-Cas9 and established the framework for its genome-editing applications [41]. Below, we reconstruct the key experimental approaches and methodologies.
In Vitro Reconstitution of Cas9 Complex
Objective: To demonstrate that purified Cas9 protein complexed with guide RNA can cleave target DNA in vitro in a programmable manner.
Reagents and Materials:
- Purified Streptococcus pyogenes Cas9 protein
- Synthetic crRNAs with varying spacer sequences
- In vitro transcribed tracrRNA
- Target DNA plasmids containing matching protospacer sequences
- Control plasmids with mismatched targets
Protocol:
- Complex Formation: Incubate Cas9 protein (100 nM) with crRNA (120 nM) and tracrRNA (120 nM) in reaction buffer (20 mM HEPES pH 7.5, 150 mM KCl, 1 mM MgCl₂, 10% glycerol) for 15 minutes at 37°C [41].
- DNA Cleavage Reaction: Add target DNA plasmid (10 nM) to the pre-formed Cas9-RNA complex in cleavage buffer (20 mM HEPES pH 7.5, 100 mM KCl, 5 mM MgCl₂, 1 mM DTT).
- Time Course Incubation: Aliquot reactions at 0, 5, 15, 30, and 60 minutes.
- Reaction Termination: Add STOP buffer (50 mM EDTA, 1.2% SDS).
- Analysis: Separate cleavage products by agarose gel electrophoresis and visualize with ethidium bromide staining.
Key Results: The experiment demonstrated sequence-specific DNA cleavage dependent on both Cas9 protein and guide RNA components. Cleavage efficiency reached >90% within 60 minutes and required complementarity between the crRNA spacer and target DNA [41].
sgRNA Engineering and Validation
Objective: To simplify the two-RNA system (crRNA + tracrRNA) by creating a single-guide RNA (sgRNA) chimera.
Design Strategy: The sgRNA was designed by connecting the 3' end of the crRNA spacer to the 5' end of tracrRNA via a synthetic GAAA tetraloop, preserving the secondary structure required for Cas9 binding and activation [41].
Validation Experiment:
- In Vitro Transcription: Synthesize sgRNA with T7 RNA polymerase.
- Cleavage Assay: Compare DNA cleavage efficiency between the natural two-RNA system and the engineered sgRNA system.
- Specificity Testing: Test cleavage against targets with single and multiple mismatches to determine specificity.
Results: The chimeric sgRNA mediated efficient target cleavage with activity comparable to the natural two-RNA system, while significantly simplifying the experimental setup [41]. This engineering breakthrough made CRISPR-Cas9 accessible to laboratories worldwide.
PAM Specificity Analysis
Objective: To characterize the protospacer adjacent motif (PAM) requirements for Cas9 targeting.
Methodology:
- PAM Library Construction: Generate a target plasmid library with randomized 5'-NNN-3' sequences adjacent to the protospacer.
- Positive Selection: Perform Cas9 cleavage and transform into bacteria with selection for uncut plasmids.
- Sequencing Analysis: Sequence surviving plasmids to identify PAM sequences that failed to support cleavage.
Results: The experiment confirmed that SpCas9 requires a 5'-NGG-3' PAM sequence immediately following the target protospacer, with cleavage occurring 3 bp upstream of the PAM [41] [44].
Table 2: Quantitative Analysis of CRISPR-Cas9 Cleavage Efficiency
| Target Sequence | PAM | Cleavage Efficiency | Time to 50% Cleavage | Mismatch Tolerance |
|---|---|---|---|---|
| Perfect Match | NGG | >90% | 15 min | 0 mismatches |
| Single Mismatch | NGG | 45-75% | 30-45 min | Position-dependent |
| Double Mismatch | NGG | 10-40% | >60 min | Severe reduction |
| Non-canonical PAM | NAG | 15-25% | >60 min | Inefficient cleavage |
| No PAM | N/A | <1% | N/A | No cleavage |
The Researcher's Toolkit: Essential Reagents and Materials
Successful implementation of CRISPR-Cas9 genome editing requires careful selection of components and optimization of delivery methods. The table below details key research reagents and their applications.
Table 3: Essential Research Reagents for CRISPR-Cas9 Experiments
| Reagent Category | Specific Examples | Function | Considerations |
|---|---|---|---|
| Cas9 Expression Systems | SpCas9, SaCas9, eSpCas9(1.1) | DNA cleavage effector | Size, PAM specificity, fidelity |
| Guide RNA Backbones | U6-promoter driven sgRNA | Target recognition | Promoter compatibility, stability |
| Delivery Vectors | AAV, lentivirus, plasmid | Component delivery | Cargo size, tropism, efficiency |
| Repair Templates | ssODN, dsDNA donor | HDR-mediated editing | Length, symmetry, modification |
| Detection Assays | T7E1, TIDE, NGS | Editing efficiency | Sensitivity, quantitative accuracy |
| Cell Lines | HEK293, iPSCs, primary cells | Experimental context | Division rate, transfection efficiency |
Delivery Strategies for CRISPR Components
Effective delivery remains a critical challenge for CRISPR-Cas9 applications. The choice between viral and non-viral delivery methods depends on the experimental context and target cells:
Viral Vectors: Adenoviral vectors (AV) and adeno-associated viruses (AAV) offer high transduction efficiency but face packaging size limitations (~4.7 kb for AAV) that necessitate the use of smaller Cas9 orthologs like Staphylococcus aureus Cas9 (SaCas9) [44].
Non-Viral Delivery: Lipid nanoparticles (LNPs) and electroporation enable transient delivery of Cas9-gRNA ribonucleoprotein (RNP) complexes, reducing off-target effects and immune responses [44]. Recent clinical trials have demonstrated the efficacy of LNP-mediated CRISPR delivery for treating genetic disorders [44].
The groundbreaking work characterizing CRISPR-Cas9 has fundamentally transformed genetic engineering, enabling precise genome manipulation across diverse biological systems. The key insight that the bacterial immune system could be reduced to a two-component programmable platform created a versatile tool that combines unprecedented simplicity with powerful editing capabilities.
While the original CRISPR-Cas9 system continues to be widely adopted, recent advances have further expanded its capabilities. Base editors and prime editors now enable precise nucleotide changes without double-strand breaks, addressing key limitations in safety and efficiency [44] [45]. Meanwhile, AI-driven protein design has generated novel CRISPR effectors with improved properties, such as the recently described OpenCRISPR-1, which exhibits comparable activity to SpCas9 while being 400 mutations distant from natural sequences [46].
The journey from fundamental bacterial immunity research to programmable genome editing exemplifies how curiosity-driven science can yield transformative technologies. As CRISPR-based therapies enter clinical practice for conditions like sickle cell anemia and continue to expand into new therapeutic areas, the foundational work of Charpentier, Doudna, and their colleagues stands as a testament to the power of basic scientific research to revolutionize medicine and biology.
CRISPR in Action: Editing Strategies, Delivery Systems, and Clinical Trial Progress in 2025
The advent of CRISPR-Cas9 technology has instigated a profound transformation in therapeutic development, offering unprecedented capabilities for precise genetic modifications. This revolutionary gene-editing tool, derived from a natural bacterial immune system, enables researchers to permanently correct deleterious base mutations or disrupt disease-causing genes with great precision and efficiency [1]. The CRISPR-Cas9 system operates through a simplified mechanism where a guide RNA (gRNA) sequence directs the Cas9 nuclease to a specific target DNA sequence, creating a double-strand break (DSB) that harnesses the cell's innate repair mechanisms to achieve the desired genetic alteration [47] [48]. This fundamental breakthrough has opened new frontiers in treating both genetic and acquired diseases, with therapeutic applications now advancing through clinical trials and into approved medicines [1] [49].
The strategic decision between ex vivo and in vivo editing approaches represents a critical juncture in therapeutic development pipelines. Ex vivo editing involves harvesting cells from a patient, genetically modifying them outside the body, and then reinfusing the edited cells back into the patient [47]. In contrast, in vivo editing delivers the CRISPR components directly into the patient's body to edit cells within their native physiological context [47] [49]. This technical guide examines the core considerations, methodologies, and future directions for these distinct therapeutic paradigms within the framework of CRISPR-Cas9 genome editing research, providing drug development professionals with a comprehensive resource for strategic decision-making.
Fundamental Mechanisms of CRISPR-Cas9 Editing
The CRISPR-Cas9 system functions through a sophisticated yet programmable molecular mechanism that mimics adaptive immune defense in bacteria. When infected with viruses, bacteria capture small pieces of viral DNA and insert them into their own genome as CRISPR arrays, providing a genetic "memory" of past infections [48]. Upon subsequent viral attacks, bacteria transcribe these arrays into RNA segments that guide Cas proteins to recognize and cleave invading viral DNA [48]. Researchers have repurposed this natural system for precise genome engineering by creating synthetic guide RNAs that direct the Cas9 nuclease to specific genomic loci of interest [47] [1].
The CRISPR-Cas9 complex identifies specific protospacer adjacent motif (PAM) sequences adjacent to the target DNA site, inducing double-strand breaks (DSBs) in the genome [49]. Following DSB formation, eukaryotic cells activate primarily two DNA repair pathways: non-homologous end joining (NHEJ) and homology-directed repair (HDR) [47] [49]. NHEJ frequently results in small insertions or deletions (indels) at the cleavage site, often disrupting gene function and creating knockouts [47]. HDR enables precise genetic modifications using a homologous DNA template, allowing for gene correction or knock-in strategies [47] [49]. The system's core components include the Cas9 nuclease and a single-guide RNA (sgRNA) that combines the functions of CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA) into a single transcript of approximately 100 nucleotides [50] [49].
Figure 1: Fundamental Mechanism of the CRISPR-Cas9 System. The process initiates with Protospacer Adjacent Motif (PAM) sequence recognition, followed by guide RNA binding to complementary DNA sequences. The Cas9 nuclease then creates a double-strand break (DSB), which is repaired via either non-homologous end joining (NHEJ) or homology-directed repair (HDR) pathways [47] [49] [48].
Beyond standard CRISPR-Cas9 cleavage, advanced precision genome editing tools have been developed that bypass the reliance on DSBs. Base editors facilitate direct chemical conversion of one DNA base to another without breaking the DNA backbone—cytidine base editors (CBEs) convert cytosine to thymine, while adenine base editors (ABEs) convert adenine to guanine [49]. Prime editors represent an even more versatile platform, consisting of a Cas9 nickase fused to a reverse transcriptase that allows targeted incorporation of precise edits using a prime editing guide RNA (pegRNA) template [49]. These sophisticated tools significantly expand the therapeutic applications of CRISPR technology while potentially reducing off-target effects associated with traditional DSB-based approaches.
Ex Vivo Gene Editing: Principles and Applications
Core Methodology and Workflow
Ex vivo gene editing represents a well-established therapeutic paradigm wherein patient cells are harvested, genetically modified outside the body, and then reintroduced into the patient [47]. This approach begins with the collection of specific cell populations, most commonly hematopoietic stem cells (HSCs) or immune cells, through apheresis or bone marrow aspiration. The isolated cells are then activated and cultured ex vivo to make them receptive to genetic modification. The CRISPR-Cas9 components—typically delivered as ribonucleoprotein (RNP) complexes via electroporation—introduce precise genetic changes [50] [51]. Following quality control validation to confirm editing efficiency and safety, the modified cells are expanded in culture and subsequently infused back into the patient, who often undergoes conditioning chemotherapy to create space for the engineered cells [47].
The ex vivo approach offers several distinct advantages, including precise control over editing efficiency, the ability to perform thorough quality assessment before administration, and reduced concerns about immune responses to bacterial Cas proteins [47]. Additionally, ex vivo editing facilitates complex multi-step genetic engineering strategies that would be challenging to accomplish in vivo, such as the generation of chimeric antigen receptor (CAR) T-cells for immunotherapy applications [47] [52]. However, this methodology also presents significant challenges, including the technical complexity and high cost of manufacturing, potential loss of cell viability or function during the multi-step process, and the necessity for patient conditioning regimens that carry their own risks and side effects [47].
Exemplary Protocol: Ex Vivo Editing of Hematopoietic Stem Cells
The groundbreaking therapy Casgevy (exagamglogene autotemcel) for sickle cell disease and transfusion-dependent beta-thalassemia exemplifies a robust ex vivo editing protocol [47]. The following detailed methodology outlines the key experimental procedures:
HSC Collection and Isolation: Hematopoietic stem and progenitor cells are harvested from the patient via apheresis, followed by CD34+ cell selection using immunomagnetic beads to enrich the target population [47].
Cell Activation and Culture: Isolated CD34+ cells are activated in serum-free media supplemented with cytokines (SCF, TPO, FLT3-L) for 24-48 hours to promote cell cycle entry, which enhances CRISPR editing efficiency [47].
CRISPR Component Delivery: Activated cells are electroporated using a specialized system (e.g., Lonza 4D-Nucleofector) with precomplexed Cas9-gRNA ribonucleoprotein (RNP) targeting the BCL11A erythroid-specific enhancer region. The RNP complex is formed by incubating purified Cas9 protein with synthetic sgRNA at a molar ratio of 1:2 for 10-20 minutes at room temperature prior to electroporation [47].
Post-Editing Culture and Expansion: Edited cells are cultured in expansion media for 2-3 days, allowing expression of the modified genetic program and monitoring editing efficiency via PCR-based assays and Sanger sequencing [47].
Patient Conditioning and Reinfusion: Patients receive myeloablative conditioning with busulfan to create marrow niche space, followed by intravenous infusion of the cryopreserved, edited CD34+ cells. Patients are monitored for engraftment and hematopoietic recovery [47].
In the pivotal clinical trials (CLIMB-111, CLIMB-121, and CLIMB-131), this ex vivo approach demonstrated remarkable efficacy. For sickle cell disease patients, the treatment resulted in a profound reduction of vaso-occlusive crises, with 59% (26 of 44) of patients experiencing complete resolution of major adverse events [47]. The editing strategy disrupts an enhancer region of the BCL11A gene, leading to sustained reactivation of fetal hemoglobin production, which compensates for the defective adult hemoglobin in these hemoglobinopathies [47] [49].
Figure 2: Ex Vivo Gene Editing Workflow. This schematic illustrates the multi-step process of ex vivo gene editing, from initial cell collection from the patient through genetic modification outside the body to final reinfusion of edited cells [47].
In Vivo Gene Editing: Principles and Applications
Core Methodology and Workflow
In vivo gene editing represents a more direct therapeutic approach wherein CRISPR-Cas9 components are delivered systemically or locally to edit cells within the patient's own body [47] [49]. This strategy requires sophisticated delivery vehicles to transport the large CRISPR machinery to target tissues while avoiding degradation, immune recognition, and off-target effects. The primary delivery modalities include viral vectors (particularly adeno-associated viruses - AAVs), lipid nanoparticles (LNPs), and other non-viral vectors [50] [49]. Each delivery system presents distinct advantages and limitations related to packaging capacity, tropism, immunogenicity, and manufacturing scalability.
The in vivo approach offers several compelling advantages, including simplified treatment administration that may enable broader clinical accessibility, the potential to target tissues and cell types that cannot be easily removed or manipulated ex vivo, and the ability to address neurological, muscular, and retinal disorders directly at the disease site [9] [49]. Furthermore, LNP-mediated in vivo delivery enables the possibility of redosing, as demonstrated in recent clinical cases where patients safely received multiple treatments to increase editing efficiency—an option generally not feasible with viral vectors due to immune responses [9]. However, significant challenges remain, including potential immune reactions to bacterial Cas proteins or delivery vehicles, difficulties in achieving efficient editing in therapeutically relevant cell populations, and greater concerns about off-target effects due to less control over editing distribution [50] [49].
Exemplary Protocol: LNP-Mediated In Vivo Liver Editing
Intellia Therapeutics' phase I trial for hereditary transthyretin amyloidosis (hATTR) exemplifies a robust in vivo editing protocol using LNP delivery [9]. The following detailed methodology outlines the key experimental procedures:
CRISPR Formulation: Cas9 mRNA and sgRNA targeting the TTR gene are encapsulated in liver-tropic LNPs composed of ionizable lipids, phospholipids, cholesterol, and PEG-lipid using microfluidic mixing technology. The formulated LNPs typically have a particle size of 70-100 nm and are stored at -80°C until administration [9].
Quality Control Validation: LNP formulations undergo comprehensive characterization, including encapsulation efficiency measurement (typically >90%), endotoxin testing, sterility validation, and potency assays in relevant cell lines [9].
Dose Administration: Patients receive a single intravenous infusion of LNP-formulated CRISPR components over 2-4 hours, with dosage based on body weight (ranging from 0.1-1.0 mg/kg in phase I trials). Premedication with antihistamines and corticosteroids may be administered to minimize infusion reactions [9].
Pharmacodynamic Monitoring: Blood samples are collected at regular intervals to quantify reduction in serum TTR protein levels using immunoassays, serving as a direct biomarker of editing efficacy. Additional monitoring includes assessment of liver enzymes and inflammatory markers [9].
Efficacy and Safety Follow-up: Patients are monitored for clinical improvement in neuropathy or cardiomyopathy symptoms, along with comprehensive safety assessments including immunological profiling, liver function tests, and potential off-target analysis [9].
In clinical results published in the New England Journal of Medicine, this in vivo approach demonstrated remarkable efficacy, with participants showing an average of approximately 90% reduction in levels of the disease-related TTR protein sustained throughout the length of the trial [9]. All 27 participants who reached two years of follow-up showed a sustained response with no evidence of the effect weakening over time, supporting the potential of in vivo editing for durable therapeutic effects [9].
rAAV Vector Innovations for In Vivo Delivery
Recombinant adeno-associated virus (rAAV) vectors have emerged as prominent vehicles for in vivo CRISPR delivery due to their favorable safety profile, high tissue specificity, and ability to induce sustained transgene expression [49]. However, the limited packaging capacity of rAAV vectors (<4.7 kb) presents a significant challenge for delivering CRISPR components [50] [49]. Innovative strategies have been developed to overcome this limitation:
Compact Cas Orthologs: Smaller Cas proteins such as Campylobacter jejuni Cas9 (CjCas9), Staphylococcus aureus Cas9 (SaCas9), and Cas12f enable packaging into single rAAV vectors while maintaining editing efficiency [49]. For example, subretinal delivery of rAAV8 vectors encoding CasMINI_v3.1/ge4.1 achieved transduction efficiencies of over 70% in retinal cells of disease models [49].
Dual rAAV Vector Systems: This approach splits CRISPR components across two separate rAAV vectors—one encoding the Cas nuclease and the other containing the gRNA expression cassette [49]. Co-infection of the same cell with both vectors enables reconstitution of functional editing machinery, though efficiency can be variable.
Trans-Splicing AAV Vectors: Advanced rAAV designs utilize intein-mediated protein trans-splicing to reconstitute large Cas proteins from two separate vectors, expanding the size range of proteins that can be delivered [49].
The first in vivo CRISPR-based therapy to enter human trials, EDIT-101 for Leber Congenital Amaurosis type 10 (LCA10), employs rAAV5 vectors delivered via subretinal injection to deliver SpCas9 and two gRNAs targeting intronic regions flanking the IVS26 mutation in the CEP290 gene [49]. Early findings from the phase 1/2 BRILLIANCE trial reported favorable safety outcomes and improved photoreceptor function in eleven of fourteen treated participants, supporting the feasibility of rAAV vector-mediated in vivo gene editing in humans [49].
Figure 3: In Vivo Delivery Systems for CRISPR Therapeutics. This diagram categorizes the primary delivery modalities for in vivo gene editing, highlighting key characteristics and limitations of each approach [9] [50] [49].
Direct Comparative Analysis: Strategic Considerations
Technical and Clinical Parameter Comparison
Table 1: Comprehensive Comparison of Ex Vivo vs. In Vivo Editing Approaches
| Parameter | Ex Vivo Approach | In Vivo Approach |
|---|---|---|
| Therapeutic Examples | Casgevy for sickle cell disease and beta-thalassemia [47] | EDIT-101 for LCA10 [49]; Intellia's hATTR program [9] |
| Delivery Method | Electroporation of RNP complexes [47] [51] | LNP or rAAV vectors [9] [49] |
| Editing Efficiency | High (>80% in target cell populations) [47] | Variable (0.34%-90% depending on tissue and delivery) [9] [49] |
| Manufacturing Complexity | High (cell processing, GMP facilities) [47] [51] | Moderate to high (vector production, formulation) [50] [49] |
| Treatment Regimen | Multi-step (cell collection, editing, conditioning, reinfusion) [47] | Single or limited administrations [9] |
| Dosing Strategy | One-time treatment with conditioned cells [47] | Potential for redosing (LNP-based) [9] |
| Target Tissues | Hematopoietic cells, immune cells [47] [52] | Liver, retina, CNS, muscle [9] [49] |
| Safety Monitoring | Pre-infusion quality control, engraftment monitoring [47] | Off-target assessment, immune response monitoring [50] [49] |
| Regulatory Pathway | Established for cell therapies [51] | Evolving framework for in vivo gene editing [51] |
| Commercialization Challenges | High cost, specialized treatment centers [47] [9] | Delivery optimization, immunogenicity [50] [49] |
Decision Framework for Therapeutic Development
The strategic selection between ex vivo and in vivo approaches depends on multiple interrelated factors that drug development professionals must carefully evaluate:
Target Tissue Accessibility: Tissues that can be readily harvested and manipulated (e.g., blood, bone marrow, immune cells) are strong candidates for ex vivo approaches. In contrast, tissues that cannot be easily removed or reintroduced (e.g., brain, retina, muscle) necessitate in vivo strategies [47] [49].
Disease Pathophysiology: Monogenic disorders requiring precise gene correction may favor ex vivo approaches where editing efficiency can be rigorously validated pre-administration. Diseases where gene disruption or knockdown provides therapeutic benefit (e.g., TTR amyloidosis, hypercholesterolemia) may be well-suited to in vivo approaches [9].
Therapeutic Window and Safety Profile: Ex vivo editing offers greater control over the edited product but requires patient conditioning with associated risks. In vivo editing avoids conditioning regimens but presents greater challenges in controlling distribution and potential immune responses [47] [50].
Manufacturing and Commercial Considerations: Ex vivo therapies are typically patient-specific and require complex, costly manufacturing processes. In vivo therapies have the potential for more scalable, off-the-shelf production but face delivery and targeting challenges [47] [9].
Regulatory Pathway: Ex vivo cell therapies have established regulatory precedents, while in vivo CRISPR therapies are navigating evolving regulatory frameworks with unique considerations for long-term monitoring and off-target risk assessment [51].
Figure 4: Therapeutic Development Decision Framework. This flowchart outlines key considerations for selecting between ex vivo and in vivo editing approaches based on target cell accessibility, editing requirements, and therapeutic goals [47] [9] [49].
The Scientist's Toolkit: Essential Research Reagents
Table 2: Key Research Reagents for CRISPR Therapeutic Development
| Reagent/Category | Function | Technical Considerations |
|---|---|---|
| GMP-grade Cas9 Nuclease | Protein component for clinical-grade therapeutic editing [51] | Requires stringent quality control, endotoxin testing, and activity validation [51] |
| GMP-grade Guide RNA | Targeting component for clinical applications [51] | Must be manufactured under cGMP conditions with purity >95% and minimal contaminants [51] |
| Electroporation Systems | Delivery method for ex vivo RNP transfection [50] | Optimization required for cell type-specific parameters (voltage, pulse length) [50] |
| Lipid Nanoparticles (LNPs) | Non-viral delivery vehicle for in vivo applications [9] [49] | Composition affects tropism, efficiency, and immunogenicity; liver-tropic formulations well-established [9] |
| rAAV Vectors | Viral delivery vehicle for in vivo applications [49] | Serotype selection critical for tissue tropism; packaging capacity limitation must be addressed [49] |
| Cell Separation Media | Isolation of target cell populations (e.g., CD34+ cells) [47] | Density gradient centrifugation or immunomagnetic selection for target cell enrichment [47] |
| Cytokine Cocktails | Cell activation and expansion pre-/post-editing [47] | Composition and timing critical for maintaining stemness or promoting differentiation [47] |
| Quality Control Assays | Safety and efficacy assessment [51] | Includes sterility, mycoplasma, endotoxin testing, editing efficiency, and viability assays [51] |
Future Perspectives and Emerging Innovations
The field of therapeutic genome editing continues to evolve at a remarkable pace, with several emerging innovations poised to address current limitations. Delivery technologies represent a primary focus, with ongoing development of novel nanoparticle formulations with enhanced tissue specificity beyond the liver, including brain, muscle, and lung tropism [9] [52]. The successful demonstration of LNP-mediated in vivo editing enabling multiple doses represents a significant advancement, overcoming a key limitation of viral vector approaches [9]. Additionally, the emergence of compact CRISPR systems including Cas12f, IscB, and TnpB with molecular sizes below 4 kb offers enhanced compatibility with viral vector packaging constraints and potentially reduced immunogenicity [49].
Innovative therapeutic strategies are also expanding the potential applications of both ex vivo and in vivo editing. The recent development of "disease-agnostic" approaches that could treat many patients regardless of their specific mutation addresses a critical challenge in rare disease drug development [53]. Rather than creating personalized edits for each mutation, these strategies aim for standardized methods applicable across multiple diseases, potentially improving efficiency and reducing costs [53]. In the regenerative medicine space, allogeneic, gene-edited, stem cell-derived therapies are advancing, with clinical trials underway for Type 1 diabetes using hypoimmune edits to avoid rejection without chronic immunosuppression [52].
The regulatory landscape for CRISPR therapies continues to mature, with evolving frameworks for both ex vivo and in vivo approaches [51]. The first regulatory approvals of CRISPR-based medicines have established important precedents, while the rapid development and approval of personalized in vivo therapies for ultra-rare diseases has demonstrated regulatory flexibility for addressing urgent unmet medical needs [9]. However, significant challenges remain, including standardization of potency assays, long-term monitoring protocols, and analytical methods for assessing off-target effects [51]. As the field advances, interdisciplinary integration of artificial intelligence and machine learning is expected to enhance gRNA design specificity, predict editing outcomes, and optimize delivery systems, further refining the precision and safety of therapeutic genome editing [50].
The strategic selection between ex vivo and in vivo editing approaches represents a fundamental consideration in CRISPR-based therapeutic development, with significant implications for research direction, manufacturing strategy, regulatory pathway, and ultimate clinical implementation. Ex vivo editing offers greater control over the editing process and established clinical success for hematopoietic and immune disorders, while in vivo editing provides direct access to challenging target tissues and potential for simplified treatment regimens. The remarkable clinical outcomes demonstrated by both approaches—from the transformational benefits of Casgevy for hemoglobinopathies to the profound protein reduction achieved in hATTR amyloidosis—underscore the therapeutic potential of CRISPR technologies across diverse disease contexts.
As the field continues to advance, the development of more sophisticated delivery systems, precision editing tools, and disease-agnostic strategies promises to expand the addressable patient populations and therapeutic indications. Drug development professionals must navigate the complex technical, manufacturing, and regulatory considerations specific to each approach while remaining attentive to the rapid pace of innovation in this transformative field. Through continued refinement of both ex vivo and in vivo platforms, CRISPR-based therapies are poised to deliver on the promise of precision genetic medicine, offering durable and potentially curative treatments for previously intractable diseases.
The CRISPR-Cas9 system has revolutionized genome editing by providing an unprecedented ability to modify DNA sequences with precision. However, its therapeutic and research application is critically dependent on efficient delivery into target cells. Viral vectors, particularly adeno-associated virus (AAV) and lentivirus, have emerged as leading vehicles for this purpose, each with distinct advantages and limitations that determine their suitability for specific experimental or therapeutic contexts. Within the basic principles of CRISPR-Cas9 research, understanding these delivery systems is paramount for designing effective gene editing strategies. This guide examines the technical constraints of AAV vectors, explores the complementary applications of lentiviral systems, and provides practical methodologies for researchers navigating viral vector selection for CRISPR delivery.
Core Challenges in AAV-Mediated CRISPR Delivery
The AAV Packaging Limit and Cas9 Size Constraints
The most significant limitation of AAV for CRISPR-Cas9 delivery is its constrained packaging capacity of approximately 4.7 kilobases (kb) [54]. This poses a substantial challenge for delivering the commonly used Streptococcus pyogenes Cas9 (SpCas9), which, together with its guide RNA (gRNA) and necessary regulatory elements, requires about 4.2 kb [54]. This leaves minimal space for promoters, enhancers, or other regulatory sequences, often necessitating creative engineering solutions for effective delivery.
Table 1: Strategies to Overcome AAV Packaging Limitations
| Strategy | Mechanism | Examples | Key Considerations |
|---|---|---|---|
| Smaller Cas Orthologs | Utilize naturally compact Cas9 proteins | SaCas9 (3.2 kb), CjCas9 (3.0 kb), Nme2Cas9 (3.2 kb) [54] | PAM specificity may limit targetable genomic sites; efficiency varies |
| Split Intein Systems | Divide Cas9 into two fragments packaged into separate AAVs; reconstituted via protein trans-splicing [54] | Split SpCas9 | Reconstitution efficiency can be modest compared to full-length Cas9 |
| Dual AAV Vectors | Separate Cas9 and gRNA expression cassettes into two individual AAV particles [54] | AAV-SpCas9 + AAV-gRNA | Requires high co-infection rate; increases manufacturing complexity |
Immunogenicity and Safety Profile
While AAV vectors are generally favored for their low immunogenicity and non-pathogenic nature [55], concerns remain regarding immune responses against both the AAV capsid and the delivered Cas9 transgene. Pre-existing immunity to common AAV serotypes in human populations can neutralize the vector before it reaches target cells, reducing efficacy [56]. Furthermore, Cas9 expression can elicit cell-mediated immune responses that may clear edited cells [56]. Although AAV predominantly exists as episomal DNA, minimizing genotoxicity risks, the potential for rare genomic integration events and immunogenic reactions necessitates careful safety profiling.
Lentiviral Vector Applications for CRISPR Delivery
Advantages for Complex Editing Scenarios
Lentiviral vectors (LVs) offer a complementary set of advantages that make them ideal for many CRISPR applications, particularly where AAV constraints are prohibitive. Their most significant advantage is a larger packaging capacity of 8-12 kb [57], which readily accommodates SpCas9, multiple gRNAs, and complex regulatory elements within a single vector. LVs facilitate stable integration into the host genome, enabling long-term, persistent expression of CRISPR components in both dividing and non-dividing cells [58] [57]. This is particularly valuable for pooled screening applications and generating stable knockout cell lines.
Safety Engineering in Lentiviral Systems
To address safety concerns related to viral integration, modern lentiviral systems incorporate several key engineering features. Self-inactivating (SIN) designs delete the enhancer/promoter region from the 3' LTR, which is copied to the 5' LTR during reverse transcription, rendering the integrated provirus transcriptionally inactive and reducing the risk of insertional mutagenesis [58]. Furthermore, third-generation packaging systems split viral genes across multiple plasmids, significantly minimizing the chance of generating replication-competent lentiviruses (RCL) through recombination [58]. For applications where even transient integration is undesirable, integrase-deficient lentiviral vectors (IDLVs) provide episomal persistence with reduced genotoxic risk [58].
Table 2: AAV vs. Lentiviral Vectors for CRISPR Delivery
| Parameter | Adeno-Associated Virus (AAV) | Lentivirus |
|---|---|---|
| Packaging Capacity | ~4.7 kb [54] [57] | 8-12 kb [57] |
| Genomic Integration | Predominantly episomal (non-integrating) [57] [59] | Stable integration into host genome [58] [57] |
| Expression Kinetics | Rapid onset, typically transient (weeks-months) | Delayed onset, long-term stable |
| Typical Applications | In vivo gene editing, high-transient expression, clinical therapies [55] [59] | Stable cell line generation, pooled screens, hard-to-transfect cells [60] [61] |
| Key Advantage | High transduction efficiency in vivo, favorable safety profile [55] | Large cargo capacity, stable expression in dividing cells [58] |
| Primary Limitation | Small packaging size, pre-existing immunity [54] [56] | Risk of insertional mutagenesis, more complex production [58] |
Experimental Protocols for Viral CRISPR Delivery
Protocol: AAV-Mediated In Vivo Gene Knockout Using SaCas9
This protocol outlines the key steps for using the compact SaCas9 for in vivo gene editing, as demonstrated in mouse liver [54].
Vector Design and Packaging:
- Clone SaCas9 (3.2 kb) and a target-specific sgRNA expression cassette into an AAV transfer plasmid, flanked by AAV2 ITRs.
- Select a tissue-specific promoter (e.g., liver-specific TBG promoter) and a U6 promoter for sgRNA expression.
- Package the recombinant genome into an appropriate serotype capsid (e.g., AAV8 for liver tropism) using a triple-transfection system in HEK293 cells.
Purification and Titration:
- Purify AAV vectors from cell lysates via iodixanol gradient ultracentrifugation.
- Treat with Benzonase to degrade unpackaged DNA.
- Determine vector genome (vg) titer using quantitative PCR. A standard research dose ranges from (1 \times 10^{11}) to (1 \times 10^{12}) vg per mouse.
In Vivo Delivery and Analysis:
- Administer the AAV-SaCas9-sgRNA vector via systemic injection (e.g., tail vein) into adult mice.
- After 1-4 weeks, harvest the target tissue (e.g., liver).
- Assess editing efficiency by extracting genomic DNA and using the T7 Endonuclease I (T7E1) assay or next-generation sequencing to quantify indel formation at the target locus.
- Monitor phenotypic changes and tissue histology to confirm functional knockout and absence of toxicity.
Protocol: Generating Stable Knockout Cells with All-in-One Lentiviral CRISPR Vectors
This protocol describes the use of single-vector lentiviral systems to create stable Cas9-expressing cell pools for consistent gene editing [60] [61].
Virus Production:
- Obtain an all-in-one lentiviral transfer plasmid expressing SpCas9, a puromycin resistance gene (via a P2A or T2A peptide), and a cloning site for a target-specific gRNA under a U6 promoter.
- Co-transfect this plasmid with second-generation (psPAX2) and envelope (pMD2.G - VSV-G) packaging plasmids into HEK293T cells using a standard transfection reagent.
- Collect virus-containing supernatant at 48 and 72 hours post-transfection.
- Concentrate the virus by ultracentrifugation and determine functional titer (Transducing Units/mL, TU/mL) on HEK293T cells.
Cell Transduction and Selection:
- Transduce the target cells with the lentiviral supernatant in the presence of a transduction enhancer like polybrene (e.g., 8 µg/mL).
- 48 hours post-transduction, begin selection with the appropriate antibiotic (e.g., 1-5 µg/mL puromycin) for 5-7 days to establish a stable pool of Cas9-expressing cells.
Validation of Knockout:
- For gene knockout, transduce the stable Cas9-expressing cells with a second lentivirus expressing the target-specific gRNA or use transient transfection of a gRNA plasmid.
- After 5-7 days, analyze editing efficiency via the T7E1 assay or sequencing.
- Confirm protein-level knockout by western blot or flow cytometry, if a specific antibody is available.
Visualization of Key Concepts
AAV Engineering Strategies for CRISPR Delivery
The diagram below illustrates the three primary engineering strategies used to deliver CRISPR-Cas9 using AAV vectors.
Lentiviral Workflow for Stable Cell Line Generation
This diagram outlines the key steps in using an all-in-one lentiviral CRISPR system to generate stable knockout cell lines.
The Scientist's Toolkit: Key Research Reagents
Table 3: Essential Reagents for Viral CRISPR Research
| Reagent / Material | Function in Research | Example Specifications |
|---|---|---|
| All-in-One Lentiviral CRISPR Construct | Single vector system for stable Cas9 and gRNA expression. | EF1α/CMV promoter for Cas9, U6 for gRNA, puromycin/blasticidin resistance [61]. |
| AAV Transfer Plasmid with ITRs | Backbone for cloning expression cassette into AAV. | Contains AAV2 ITRs, multiple cloning site, compact promoter (e.g., EFS) [54]. |
| Packaging Plasmids (AAV) | Provide Rep/Cap and Helper functions for AAV production. | Serotype-specific Cap plasmid (e.g., AAV8, AAV9), pHelper [57]. |
| Packaging Plasmids (Lentivirus) | Provide viral proteins for lentivirus production. | Second-gen: psPAX2 (gag/pol/rev); Third-gen: split gag/pol + rev [58] [57]. |
| VSV-G Envelope Plasmid | Pseudotypes viral particles for broad tropism. | pMD2.G is commonly used for both AAV and LV [58] [57]. |
| HEK293T Producer Cell Line | High-transfection efficiency cells for virus packaging. | SV40 T-antigen expressing variant for high plasmid replication [57]. |
| Titer Quantification Kits | Measure functional and physical particle concentration. | qPCR for vector genome titer, ELISA for capsid titer, flow cytometry for TU [57]. |
The selection between AAV and lentiviral vectors for CRISPR-Cas9 delivery is not a matter of superiority but of strategic application. AAV's favorable safety profile and high in vivo transduction efficiency make it the vector of choice for direct in vivo therapies, despite its packaging limitations, which are being actively overcome through protein engineering and split-vector approaches. Conversely, lentiviral vectors, with their large cargo capacity and stable integration, remain indispensable tools for complex editing scenarios, extensive screening campaigns, and ex vivo cell engineering. As the field of genomic medicine advances, the complementary use of both systems, tailored to the specific experimental or therapeutic goal, will continue to underpin the successful translation of CRISPR-based technologies from bench to bedside.
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) system has revolutionized genome editing by providing an unprecedented tool for precise DNA modification [50]. Derived from a bacterial adaptive immune system, this technology enables researchers to make targeted double-strand breaks in genomic DNA, facilitating gene knock-out via non-homologous end joining (NHEJ) or precise gene correction via homology-directed repair (HDR) when a donor template is present [62] [63]. The system consists of two key components: the Cas9 nuclease, which acts as molecular scissors, and a guide RNA (gRNA) that directs Cas9 to a specific genomic locus through complementary base-pairing [50] [64].
Despite its transformative potential, the clinical application of CRISPR-Cas9 faces a significant bottleneck: the safe and efficient delivery of its components into target cells [65] [66]. The large size of the Cas9 protein (~160 kDa) and the negative charge of nucleic acid components create substantial barriers to cellular uptake and nuclear localization [64] [67]. While viral vectors have been widely used for gene delivery, concerns regarding immunogenicity, insertional mutagenesis, and limited packaging capacity have motivated the development of non-viral alternatives [65] [64]. Among these, lipid nanoparticles (LNPs) and electroporation have emerged as leading platforms, each offering distinct advantages and limitations for research and therapeutic applications [68] [67].
This review examines recent breakthroughs in these two non-viral delivery methodologies, providing a technical framework for their implementation in CRISPR-Cas9 genome editing research. We present comparative performance data, detailed protocols, and practical guidance to assist researchers in selecting and optimizing delivery strategies for specific experimental contexts.
Comparative Performance Analysis of Delivery Platforms
The choice between LNP-mediated delivery and electroporation depends on multiple factors, including target cell type, application (in vivo vs. ex vivo), desired editing efficiency, and viability requirements. The tables below summarize quantitative data from recent studies to guide this decision-making process.
Table 1: Comparative Performance of LNP Formulations for CRISPR-Cas9 Delivery
| Cargo Format | Cell Type/Model | Editing Efficiency | Key Advantages | Reference |
|---|---|---|---|---|
| mRNA Cas9 + sgRNA | HEK293T (in vitro) | Higher than RNP format | Smaller particle size, better enzyme protection | [62] |
| mRNA Cas9 + sgRNA | HEPA 1-6 (in vitro) | Higher than RNP format | Superior editing efficiency | [62] |
| mRNA Cas9 + sgRNA | Ai9 mice (in vivo) | 60% knock-out in hepatocytes | Liver-specific biodistribution | [62] |
| Cas9 RNP + HDR | HEK293T (in vitro) | Lower than mRNA format | - | [62] |
| Cas9 RNP + HDR | Ai9 mice (in vivo) | Not detected | Spleen and lung biodistribution | [62] |
| CAR-mRNA | Primary T cells | Prolonged efficacy vs. electroporation | Extended mRNA persistence, less cytotoxicity | [68] |
Table 2: Electroporation Efficiency Across Cell Types
| Cell Type | Optimal Parameters | Editing Efficiency | Viability | Reference |
|---|---|---|---|---|
| SaB-1 (marine teleost) | 1800 V, 20 ms, 2 pulses | ~95% | ~20% | [69] |
| DLB-1 (marine teleost) | 1700 V, 20 ms, 2 pulses | ~28% | Reduced | [69] |
| DLB-1 (marine teleost) | 1600 V, 15 ms, 3 pulses | ~10% | ~50% | [69] |
| HSPCs (CASGEVY) | Protocol-specific | Up to 90% indels | Clinically acceptable | [67] |
Table 3: Strategic Selection Guide for Delivery Methods
| Application Context | Recommended Method | Rationale | Considerations |
|---|---|---|---|
| In vivo delivery | Lipid Nanoparticles | Superior biodistribution, organ-specific targeting | Liver accumulation predominant; formulation critical |
| Ex vivo cell engineering | Electroporation | High efficiency in hard-to-transfect cells | Viability trade-offs; requires parameter optimization |
| Primary immune cells | LNPs (mRNA) | Reduced cytotoxicity, prolonged expression | Better persistence than electroporation for CAR-T [68] |
| Marine teleost cells | Electroporation | Species-specific barriers overcome | Cell line-dependent efficiency [69] |
| Clinical translation (ex vivo) | Electroporation | Proven platform (CASGEVY) | Regulatory precedent established [67] |
Lipid Nanoparticle-Mediated Delivery
Mechanism and Advantages
Lipid nanoparticles represent a leading non-viral platform for CRISPR-Cas9 delivery, particularly for in vivo applications. LNPs function by encapsulating nucleic acid payloads within a protective lipid bilayer, shielding them from degradation and facilitating cellular uptake through endocytosis [66] [64]. Their core advantage lies in their customizable composition, which allows researchers to tailor properties such as size, surface charge, and targeting specificity to particular tissues or cell types [66].
Recent comparative studies have revealed that the choice of cargo format significantly influences editing outcomes. LNP formulations encapsulating mRNA encoding Cas9 along with sgRNA have demonstrated superior editing efficiency compared to RNP-loaded LNPs in both in vitro and in vivo models [62]. This performance advantage is attributed to several factors: smaller particle sizes, enhanced protection against degrading enzymes, and more efficient processing of the editing machinery when components are expressed intracellularly rather than delivered pre-formed [62].
LNP Formulation Protocol
The following detailed protocol for LNP formulation has been adapted from established methodologies for in vitro transfection [70]:
Calculation of Lipid Components:
- Determine the required mRNA amount (in ng)
- Calculate the N/P ratio (moles of positively charged nitrogens in ionizable lipids (N) / moles of negatively charged phosphates in mRNA (P))
- Convert mRNA mass to P value using the average molecular weight of ribonucleotides (337.45 g/mol)
- Calculate the N value based on the desired N/P ratio (typically N/P = 6-6.5 for commercial formulations)
- Calculate moles of ionizable lipids based on the N value
- Determine required moles for each lipid component based on the molar ratio (e.g., SM-102:DSPC:Cholesterol:DMG-PEG2000 = 50:10:38.5:1.5)
Preparation of Lipid Stock Solutions:
- Weigh lipids precisely using an analytical balance
- Dissolve in methanol-chloroform mixed solvent (1:1, v/v) and vortex thoroughly
- For DMG-PEG2000, use ethanol as solvent instead
- Prepare fresh solutions and avoid long-term storage
Lipid Mixture Film Formation:
- Combine 1.1× volumes of each lipid component in a 2-mL glass vial
- Evaporate organic solvents using a rotary evaporator at approximately 40°C for 5 minutes
- Store the resulting lipid film at -20°C if not used immediately
LNP Formation via Thermo-shaker:
- Redissolve the lipid film in 55 μL of ethanol with sonication and vortexing if necessary
- Prepare mRNA solution by diluting mRNA stock in citrate buffer (pH 4.0) to 153 μL
- Add 50 μL of lipid solution to a clean Eppendorf tube placed on a thermo-shaker at 25°C, 1400 rpm
- Quickly add 152 μL of mRNA solution and shake for 15 seconds
Solvent Exchange and Buffer Compatibility:
- Transfer the mRNA-LNP solution to an Amicon Ultra Centrifugal Filter pre-washed with DPBS
- Add DPBS to fill the filter unit and centrifuge at 14,000 × g for 10 minutes
- Resuspend the concentrated LNPs in complete cell culture media for in vitro applications
Critical Parameters for Optimization
Successful LNP-mediated CRISPR delivery depends on several key parameters:
- N/P Ratio: The balance between cationic lipids and anionic nucleic acids critically impacts encapsulation efficiency and cellular toxicity. Commercial mRNA-LNPs typically use an N/P ratio of approximately 6.5 [70].
- Buffer Conditions: mRNA must be diluted in citrate buffer (pH 4.0) to ensure stability during formulation, while final LNPs should be resuspended in physiologically compatible buffers like DPBS [70].
- Serum Conditions: Contrary to many transfection protocols, LNP-mediated mRNA delivery achieves highest efficiency in complete media with serum, rather than under serum-starved conditions [70].
Electroporation-Based Delivery
Mechanism and Applications
Electroporation utilizes brief electrical pulses to create transient pores in cell membranes, allowing direct passage of CRISPR components into the cytoplasm. This physical method is particularly valuable for ex vivo applications where target cells are accessible and viability constraints can be managed [69] [67]. The technique shows exceptional performance in clinical settings, as demonstrated by the FDA-approved therapy CASGEVY, which uses electroporation to introduce CRISPR components into hematopoietic stem cells ex vivo for treating sickle cell disease and β-thalassemia [67].
A key advantage of electroporation is its compatibility with various CRISPR cargo formats, including plasmid DNA, mRNA, and pre-assembled ribonucleoprotein (RNP) complexes [69]. RNP electroporation is particularly attractive due to rapid editing activity, reduced off-target effects, and transient presence in cells, minimizing immune responses and potential genomic integration [69] [67].
Electroporation Optimization Protocol
Optimizing electroporation parameters is critical for balancing editing efficiency and cell viability. The following protocol, adapted from marine teleost studies with broad applicability, provides a systematic approach [69]:
Parameter Screening:
- Test a voltage range (e.g., 1500-1800 V for specific systems)
- Evaluate pulse duration (e.g., 10-20 ms)
- Assess pulse number (e.g., 1-3 pulses)
- Use Cas9 conjugated with a fluorescent dye (e.g., Cas9-Cy3) to quantify uptake efficiency
- Measure cell viability simultaneously using trypan blue exclusion or similar methods
Cell-Type Specific Optimization:
- SaB-1 cells: 1800 V, 20 ms, 2 pulses for maximum efficiency (~95% editing) despite lower viability (~20%)
- DLB-1 cells: 1600 V, 15 ms, 3 pulses for balanced efficiency (~10%) and viability (~50%)
- Alternative DLB-1 parameters: 1700 V, 20 ms, 2 pulses for higher editing (~28%) with reduced viability
RNP Complex Preparation:
- Combine purified Cas9 protein with sgRNA at appropriate molar ratios (typically 2-3 μM final concentration)
- Allow complex formation for 10-15 minutes at room temperature before electroporation
- Consider sgRNA modifications: chemically synthesized sgRNAs (e.g., from Synthego) may offer advantages over in vitro transcribed versions
Post-Electroporation Handling:
- Immediately transfer cells to pre-warmed complete medium after pulsing
- Allow recovery for 24-48 hours before assessing editing outcomes
- Use specialized media formulations for sensitive primary cells
Critical Parameters for Optimization
- Cell Line Variability: Electroporation efficiency is highly cell-type dependent. SaB-1 cells showed ~95% editing efficiency under optimal conditions, while DLB-1 cells reached only ~28% under similar parameters [69].
- Viability-Efficiency Tradeoff: Higher voltage parameters generally increase editing efficiency but reduce cell viability. The optimal balance must be determined for each application [69].
- Genomic Considerations: Some cell lines (e.g., DLB-1) may exhibit locus-specific genomic rearrangements after editing, requiring careful assessment of target integrity [69].
The Scientist's Toolkit: Essential Research Reagents
Table 4: Key Reagents for Non-Viral CRISPR Delivery Research
| Reagent/Category | Specific Examples | Function | Application Notes |
|---|---|---|---|
| Ionizable Lipids | SM-102 | Structural component for nucleic acid complexation | Core component of modern LNP formulations [70] |
| Helper Lipids | DSPC, Cholesterol | Enhance membrane stability and fusion | Improve LNP stability and endosomal escape [70] |
| PEGylated Lipids | DMG-PEG2000 | Provides stealth properties, reduces aggregation | Critical for controlling LNP size and circulation time [70] |
| Cas9 Protein | Cas9-Cy3, Cas9-FLUOGREEN | Fluorescently labeled for tracking intracellular delivery | Enables quantification of uptake efficiency [69] |
| sgRNA Formats | IVT sgRNA, Chemically modified (Synthego) | Guides Cas9 to specific genomic targets | Chemical modifications can enhance stability and editing [69] |
| Electroporation Systems | Commercial electroporators | Apply controlled electrical pulses for delivery | Parameter optimization required for each cell type [69] |
| Cell Culture Media | Complete media with FBS | Supports cell growth and maintenance | Superior to serum-free conditions for LNP transfection [70] |
The parallel development of lipid nanoparticle and electroporation technologies has substantially advanced the field of CRISPR-Cas9 genome editing by providing effective non-viral delivery strategies. LNP platforms offer particular promise for in vivo therapeutic applications due to their favorable biodistribution profiles and ability to be targeted to specific tissues [62] [66]. Meanwhile, electroporation remains the gold standard for ex vivo cell engineering applications, as demonstrated by its successful clinical implementation in CASGEVY [67].
Future directions in this field include the development of novel ionizable lipids with improved tissue specificity, the integration of stimuli-responsive elements for controlled release, and the application of artificial intelligence to optimize LNP formulation and electroporation parameters [66] [64]. As these technologies continue to mature, they will undoubtedly expand the therapeutic potential of CRISPR-Cas9 genome editing, enabling treatments for an increasingly broad spectrum of genetic disorders.
For researchers selecting between these platforms, the decision framework should consider target cell type, application context (in vivo vs. ex vivo), desired editing kinetics, and viability requirements. By applying the optimized protocols and comparative data presented herein, scientists can effectively leverage these powerful non-viral delivery systems to advance their genome editing research.
The discovery of the CRISPR-Cas9 system has revolutionized genetic engineering, providing researchers with an unprecedented ability to modify genomes. This bacterial adaptive immune system has been repurposed as a programmable gene-editing tool that enables precise modifications at targeted genomic locations [1]. The fundamental CRISPR-Cas9 machinery consists of two key components: the Cas9 nuclease, which acts as a "molecular scissor" to cut DNA, and a guide RNA (gRNA), which directs Cas9 to a specific DNA sequence complementary to the gRNA [50]. The system operates through three stages: adaptation, expression, and interference, ultimately generating double-strand breaks (DSBs) at targeted sites [50].
While powerful, the native CRISPR-Cas9 system has significant limitations for therapeutic applications. The creation of DSBs triggers cellular DNA repair mechanisms, primarily non-homologous end joining (NHEJ) or homology-directed repair (HDR). NHEJ is error-prone and often results in insertions or deletions (indels), while HDR, though precise, requires a DNA repair template and is inefficient in non-dividing cells [71] [50]. Furthermore, DSBs can lead to unintended genomic rearrangements, chromosomal abnormalities, and activation of DNA damage response pathways that may cause cellular toxicity or apoptosis [72] [71]. These limitations prompted the development of more precise editing technologies that could overcome the reliance on DSBs while expanding the repertoire of possible genetic modifications.
Base Editing: Precision Chemical Surgery for Single Nucleotides
Fundamental Principles and Mechanism
Base editing represents a significant advancement in precision genome engineering by enabling direct chemical conversion of one DNA base to another without creating DSBs. First described in 2016, base editors utilize a catalytically impaired Cas9 variant (dCas9) fused to a nucleobase deaminase enzyme [73] [71]. Unlike standard CRISPR-Cas9, base editors do not cut both DNA strands; instead, they chemically alter specific nucleotides within a narrow editing window.
The base editing process involves several key steps. The dCas9-guide RNA complex localizes to the target DNA sequence, where dCas9 partially unwinds the DNA duplex. The deaminase enzyme then acts on a specific nucleotide within the exposed single-stranded DNA region, converting cytidine to uridine (C•G to T•A) or adenosine to inosine (A•T to G•C) [71]. Cellular DNA repair machinery subsequently processes these intermediate products to complete the base conversion. To minimize unintended editing, base editors often incorporate additional components such as uracil glycosylase inhibitor (UGI) proteins that prevent specific repair pathways from reversing the edits [71].
Base Editor Architectures and Applications
Two primary classes of base editors have been developed: cytosine base editors (CBEs) for C•G to T•A conversions, and adenine base editors (ABEs) for A•T to G•C conversions [71]. CBEs typically consist of a cytidine deaminase (such as APOBEC1) fused to dCas9 along with UGI proteins, while ABEs utilize engineered tRNA adenosine deaminase (TadA) variants fused to dCas9 [71].
Base editors offer several advantages over conventional CRISPR-Cas9 nucleases. They achieve higher efficiency of precise base changes (typically 15-75% without selection) while producing significantly fewer indels (typically <1%) [71]. This makes them particularly valuable for therapeutic applications requiring precise single-nucleotide corrections, such as correcting point mutations that cause genetic disorders. Additionally, base editors can function in non-dividing cells where HDR is inefficient, expanding their potential therapeutic utility [71].
Table 1: Comparison of Major Base Editing Systems
| Editor Type | Key Components | Base Conversion | Editing Window | Primary Applications |
|---|---|---|---|---|
| Cytosine Base Editor (CBE) | dCas9-cytidine deaminase-UGI | C•G → T•A | ~5 nucleotides | Disease modeling, pathogenic SNP correction |
| Adenine Base Editor (ABE) | dCas9-adenine deaminase | A•T → G•C | ~5 nucleotides | Therapeutic point mutation correction |
| Dual Base Editor | dCas9-cytidine/adenine deaminase | C•G → T•A + A•T → G•C | ~5 nucleotides | Simultaneous multi-base editing |
Experimental Considerations for Base Editing
When designing base editing experiments, researchers must consider several technical factors. Guide RNA design must account for the positioning of the target base within the editing window relative to the PAM sequence. The editing efficiency can vary significantly based on chromatin accessibility, sequence context, and cell type. Additionally, base editors can cause bystander edits where non-target bases within the editing window are modified, requiring careful optimization and validation [71].
Delivery of base editing components can be achieved through multiple methods. Plasmid DNA transfection works well for easily transfectable cells, while ribonucleoprotein (RNP) complexes offer reduced off-target effects and transient activity. For in vivo applications, viral vectors (AAV, lentivirus) or lipid nanoparticles (LNPs) can deliver base editor components, though size constraints of AAV (~4.7 kb) may necessitate the use of smaller Cas9 orthologs or split-intron systems [50] [9].
Prime Editing: Search-and-Replace Genome Editing
Fundamental Principles and Mechanism
Prime editing, first described in 2019, represents a more versatile precise genome editing technology that can mediate all 12 possible base-to-base conversions, as well as targeted insertions and deletions, without requiring DSBs or donor DNA templates [72] [74]. This "search-and-replace" editing system expands the capabilities of precise genome editing beyond the limitations of base editing.
The prime editing system consists of two main components: (1) a prime editor protein, which is a fusion of a Cas9 nickase (H840A) and an engineered reverse transcriptase (RT), and (2) a specialized prime editing guide RNA (pegRNA) [72] [74]. The pegRNA not only directs the complex to the target DNA site but also encodes the desired edit and contains a primer binding site (PBS) that facilitates the reverse transcription process.
The prime editing mechanism involves multiple coordinated steps. First, the pegRNA directs the prime editor to the target DNA sequence, where the Cas9 nickase nicks the non-target DNA strand. The exposed 3' end hybridizes with the PBS sequence of the pegRNA, serving as a primer for the RT. The RT then uses the RT template sequence of the pegRNA to synthesize a DNA flap containing the desired edit [72] [74]. Cellular repair processes subsequently resolve this edited flap into the genome, permanently incorporating the genetic change.
Prime Editing Mechanism: Step-by-step process of search-and-replace genome editing
Evolution of Prime Editing Systems
Since the initial development of prime editing, several generations of improved systems have been developed. The original PE1 system established the proof-of-concept but had limited efficiency. PE2 incorporated an engineered reverse transcriptase with five mutations (D200N/L603W/T330P/T306K/W313F) that enhanced stability and processivity, resulting in 2-3 fold higher editing efficiency [72] [75]. PE3 added a second nicking sgRNA to target the non-edited strand, further improving efficiency by 2-4 fold but slightly increasing indel formation [72] [75].
More recent advancements include PE4 and PE5, which incorporate dominant-negative MMR proteins (MLH1dn) to suppress mismatch repair and enhance editing efficiency [72]. The latest PE6 systems utilize compact reverse transcriptases from organisms like E. coli (Ec48) or S. pombe (Tf1) that are smaller yet maintain high efficiency, addressing delivery challenges [75]. These systems have demonstrated capability to correct mutations associated with Tay-Sachs disease in patient fibroblasts with higher efficiency than previous editors while being approximately 33% smaller in size [75].
Table 2: Evolution of Prime Editing Systems
| Editor Version | Key Improvements | Editing Efficiency | Indel Formation | Primary Applications |
|---|---|---|---|---|
| PE1 | Original proof-of-concept | ~10-20% | Low | Initial validation |
| PE2 | Engineered RT (pentamutant M-MLV) | ~20-40% | Low | Standard precise edits |
| PE3 | Additional nicking sgRNA | ~30-50% | Moderate | High-efficiency editing |
| PE4/PE5 | MMR inhibition (MLH1dn) | ~50-80% | Low | Challenging genomic contexts |
| PE6 | Compact RT variants (Ec48, Tf1) | ~70-90% | Low | Therapeutic applications with size constraints |
Experimental Considerations for Prime Editing
Successful prime editing experiments require careful pegRNA design, which must include several key elements: the spacer sequence for target recognition, the primer binding site (PBS) typically 8-15 nucleotides long, and the RT template encoding the desired edit [74]. Optimization of PBS length and RT template design significantly impacts editing efficiency. Modified pegRNAs with 3' structural motifs (e.g., evopreQ1, mpknot) can enhance stability and resistance to exonucleases, further improving outcomes [75].
The PE3 system requires an additional sgRNA to nick the non-edited strand, which should be designed to bind 40-90 bp from the pegRNA nicking site [72]. Recent variations like PE3b optimize the timing of the second nick to reduce indel formation [74]. For delivery, the large size of prime editors presents challenges for packaging into AAV vectors, necessitating the use of dual-AAV systems, non-viral delivery methods, or smaller Cas9 orthologs like Cas12a-based prime editors [72] [74].
Comparative Analysis of Editing Technologies
Technical Performance Metrics
When selecting a genome editing approach for specific applications, researchers must consider multiple performance characteristics. Conventional CRISPR-Cas9 nucleases remain the most efficient tool for gene disruption but produce unpredictable editing outcomes. Base editors offer higher precision for specific base transitions with minimal indel formation but are restricted in the types of changes they can introduce. Prime editors provide the greatest versatility in edit types while maintaining high precision but have historically shown variable efficiency across different targets and cell types [71].
Table 3: Comprehensive Comparison of Genome Editing Technologies
| Characteristic | CRISPR-Cas9 Nuclease | Base Editors | Prime Editors |
|---|---|---|---|
| DNA Break Type | Double-strand break | Single-strand break | Single-strand break |
| Editing Types | Indels, large deletions | Specific base transitions (C>T, A>G) | All base substitutions, insertions, deletions |
| Typical Efficiency | High (often >80%) | Moderate to high (15-75%) | Variable (10-50% optimized) |
| Off-target Effects | DSB-associated indels, chromosomal rearrangements | Bystander edits, off-target deamination | Minimal off-target activity |
| Product Purity | Low (mixed outcomes) | High (specific base changes) | High (precise intended edits) |
| Therapeutic Applicability | Limited by DSB toxicity | Moderate (point mutations only) | High (versatile editing capabilities) |
| Key Advantages | High efficiency for gene disruption | High precision for point mutations | Versatility without DSBs |
| Primary Limitations | Unpredictable outcomes, DSB toxicity | Restricted editing types, bystander edits | Variable efficiency, delivery challenges |
Practical Implementation Guidelines
Selection of the appropriate editing technology depends on the specific research goal. For gene knockout applications, conventional CRISPR-Cas9 remains the most straightforward approach. For specific point mutation corrections, base editors offer efficiency and precision when the required base change falls within their capabilities. For more complex edits, including transversions, small insertions, or deletions, prime editors provide the necessary versatility.
Each technology presents distinct experimental considerations. CRISPR-Cas9 requires careful analysis of editing outcomes due to heterogeneous results. Base editing experiments must account for the editing window and potential bystander effects. Prime editing requires extensive pegRNA optimization but can achieve precise edits without donor templates [72] [71].
The Scientist's Toolkit: Essential Reagents and Methodologies
Research Reagent Solutions
Successful implementation of base editing and prime editing technologies requires access to specialized reagents and tools. The following table outlines essential components for researchers entering this field.
Table 4: Essential Research Reagents for Next-Generation Editing
| Reagent Category | Specific Examples | Function | Implementation Notes |
|---|---|---|---|
| Editor Plasmids | BE4max, ABE8e, PEmax, PE6 | Encodes the editor protein | Codon-optimized versions improve expression; include selection markers |
| Guide RNA Systems | pegRNA, nicking sgRNA | Target specification and editing template | Modified scaffolds (epegRNA) improve stability; U6 promoter commonly used |
| Delivery Tools | AAV, LNPs, Electroporation | Editor component delivery | RNP delivery reduces off-targets; AAV has size constraints |
| Validation Assays | Next-generation sequencing, T7E1 | Edit confirmation and off-target assessment | Amplicon sequencing recommended for quantitative efficiency measurement |
| Cell Culture Models | HEK293T, HAP1, iPSCs | Editing experimentation | Stem cells require optimized delivery; include proper controls |
| Software Tools | pegFinder, pegRNA designer | Experimental design | In silico design improves success rate; consider PAM availability |
Protocol Framework: Prime Editing Workflow
A standardized experimental workflow for prime editing includes the following key steps:
Target Selection and pegRNA Design: Identify target site with appropriate PAM (NGG for SpCas9). Design pegRNA with 13nt PBS and 10-16nt RT template containing desired edit. Consider adding structural motifs to pegRNA 3' end for stability.
Component Delivery: Transfect cells with PE2 plasmid (or mRNA) and pegRNA plasmid. For difficult-to-transfect cells, consider RNP delivery. Include controls (untreated, pegRNA-only).
Efficiency Optimization (if needed): For challenging targets, implement PE3 system with nicking sgRNA. Test multiple PBS lengths (8-15nt) and RT template designs.
Editing Validation: Harvest cells 72-96 hours post-transfection. Extract genomic DNA and amplify target region by PCR. Sequence amplicons (Sanger or NGS) to quantify editing efficiency and byproducts.
Off-target Assessment: Perform unbiased genome-wide analysis (GUIDE-seq, CIRCLE-seq) or target potential off-target sites predicted by in silico tools.
This protocol typically requires 2-3 weeks from design to validation, with efficiency highly dependent on target site, cell type, and delivery method.
The field of precision genome editing continues to evolve rapidly. Recent advancements include the development of twin prime editing systems that can facilitate larger insertions and deletions, and CRISPR-associated transposases that enable targeted insertion of large DNA sequences without DSBs [72] [71]. The integration of artificial intelligence and machine learning approaches for editor design and optimization represents another frontier, with recent demonstrations of AI-generated editors showing comparable or improved activity and specificity relative to naturally derived systems [46].
Delivery remains a primary challenge, particularly for therapeutic applications. Ongoing research focuses on improved viral vectors with expanded packaging capacity, advanced non-viral delivery systems including lipid nanoparticles and extracellular vesicles, and cell-specific targeting strategies to enhance specificity [9] [50]. As these technologies mature, they promise to unlock new therapeutic possibilities for genetic disorders that have previously been intractable to conventional gene therapy approaches.
Base editing and prime editing technologies have fundamentally expanded the capabilities of precision genome engineering, offering researchers an increasingly sophisticated toolkit for functional genomics and therapeutic development. While each technology has distinct strengths and limitations, together they represent a powerful arsenal for addressing diverse genetic challenges. As the field continues to advance, these next-generation editors are poised to drive transformative progress across biomedical research and clinical medicine.
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) system represents a transformative technology in genome editing, derived from an adaptive immune mechanism in bacteria and archaea [76]. This prokaryotic system utilizes RNA-guided nucleases to identify and cleave specific nucleic acid sequences, a principle that has been repurposed for precise genetic manipulation across diverse organisms [77]. The simplicity, high efficiency, cost-effectiveness, and precision of CRISPR-Cas9 have positioned it as the leading genome-editing tool, surpassing earlier technologies like zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) [78] [76]. Its programmability via a short guide RNA (sgRNA) sequence complementary to target DNA enables specific genomic modifications with relative ease [76].
The clinical application of CRISPR-Cas9 has progressed rapidly from concept to reality. The first approved CRISPR-based medicine, CASGEVY (exagamglogene autotemcel), marked a historic milestone in 2023, receiving regulatory approval for sickle cell disease (SCD) and transfusion-dependent beta thalassemia (TDT) [52] [9] [79]. As of February 2025, the CRISPR clinical landscape encompasses approximately 250 clinical trials involving gene-editing therapeutic candidates across multiple disease areas, with over 150 trials currently active [80]. This whitepaper examines the technical foundations and current clinical applications of CRISPR-Cas9 across three major therapeutic domains: hematological disorders, oncology, and metabolic diseases, providing researchers with detailed experimental frameworks and resource guidance.
CRISPR-Cas9 Mechanism and Delivery Strategies
Molecular Mechanism of Action
The CRISPR-Cas9 genome editing mechanism comprises three fundamental phases: identification, cleavage, and repair [76]. The process begins with the formation of a ribonucleoprotein complex where the Cas9 nuclease associates with a synthetic single-guide RNA (sgRNA). This sgRNA directs Cas9 to a specific genomic locus through complementary base pairing between its 20-nucleotide guide sequence and the target DNA [81] [76]. A critical requirement for Cas9 activity is the presence of a protospacer adjacent motif (PAM)—a short, specific DNA sequence (5'-NGG-3' for Streptococcus pyogenes Cas9) immediately downstream of the target site [77].
Upon successful identification and binding, Cas9 induces a double-strand break (DSB) in the target DNA through its two distinct nuclease domains (HNH and RuvC) [81]. The cellular repair machinery then addresses this DSB primarily through two pathways:
- Non-Homologous End Joining (NHEJ): An error-prone repair pathway that often results in small insertions or deletions (indels) at the break site, effectively disrupting gene function and facilitating gene knockout [81] [77].
- Homology-Directed Repair (HDR): A precise repair mechanism that utilizes a donor DNA template to introduce specific genetic modifications, including point mutations or gene insertions, when sufficient template DNA is available [81].
Delivery Strategies for Clinical Applications
Effective delivery of CRISPR-Cas9 components remains a critical challenge for therapeutic applications. Current strategies can be categorized into three primary formats, each with distinct advantages and limitations for clinical translation [76]:
Viral Vector Delivery:
- Lentiviral Vectors (LV): Effectively transduce hematopoietic stem cells (HSCs) and immune cells, enabling stable genomic integration of Cas9 and sgRNA sequences.
- Adeno-Associated Viruses (AAV): Offer lower immunogenicity and non-integrating properties suitable for in vivo delivery, though limited packaging capacity necessitates split-intron systems for Cas9.
Non-Viral Delivery:
- Electroporation: Efficient for ex vivo delivery of ribonucleoprotein (RNP) complexes to primary cells including HSCs and T-cells, providing rapid editing with reduced off-target effects.
- Lipid Nanoparticles (LNPs): Emerging as the leading platform for in vivo delivery, particularly for liver-directed therapies, enabling transient expression with minimal immunogenicity [9] [82].
Physical Methods:
- Microinjection: Used primarily in research settings for zygote editing.
- Hydrodynamic Injection: Employed for preclinical liver-directed gene editing in animal models.
The choice of delivery method significantly impacts editing efficiency, specificity, and safety profile. For clinical applications, ex vivo approaches dominate hematological disorders, while in vivo strategies using LNP delivery show promise for metabolic diseases [76] [9].
Hematological Disorders
Inherited Blood Disorders
CRISPR-Cas9 has demonstrated remarkable efficacy in treating inherited hemoglobinopathies, particularly sickle cell disease (SCD) and β-thalassemia. The pioneering therapy CASGEVY utilizes ex vivo editing of autologous CD34+ hematopoietic stem and progenitor cells to disrupt the BCL11A gene, an erythroid-specific enhancer region that suppresses fetal hemoglobin (HbF) production [78] [52]. This edit induces elevated HbF levels, which compensates for defective adult hemoglobin in SCD and TDT patients, effectively eliminating vaso-occlusive crises in SCD and transfusion dependency in TDT [52] [79].
Table 1: Clinical Trials of CRISPR-Based Therapies for Hematological Disorders
| Therapy | Target | Condition | Phase | Delivery | Developer | Key Findings/Status |
|---|---|---|---|---|---|---|
| CASGEVY (exa-cel) | BCL11A enhancer | SCD, TDT | Approved (2023) | Ex vivo RNP | CRISPR Therapeutics/Vertex | >50 patients in cell collection; >50 activated treatment centers globally [52] |
| reniz-cel (EDIT-301) | BCL11A enhancer | SCD, TDT | I/II | Ex vivo (Cas12a) | Editas Medicine | Lasting efficacy and safety reported at ASH 2024 [79] |
| BEAM-101 | HBG1/2 promoters | SCD, TDT | I/II | Ex vivo base editing | Beam Therapeutics | Good efficacy and durability; potentially safer due to no DSBs [79] |
| PM359 | NCF1 | Chronic granulomatous disease | Preclinical (IND-cleared) | Ex vivo prime editing | Prime Medicine | Phase I trial predicted for early 2025 [83] |
Experimental Protocol: Ex Vivo HSC Editing for Hemoglobinopathies
Objective: Generate functional CD34+ hematopoietic stem and progenitor cells with elevated fetal hemoglobin expression through precise editing of the BCL11A erythroid enhancer region.
Materials and Reagents:
- Source Cells: Mobilized peripheral blood CD34+ cells from patient
- Editing Components: Cas9 protein, sgRNA targeting BCL11A enhancer
- Culture Media: Serum-free expansion media with cytokines (SCF, TPO, FLT3-L)
- Transfection System: Electroporation device (e.g., Lonza 4D-Nucleofector)
- QC Assays: Flow cytometry, NGS for on-target/off-target analysis, colony-forming unit assays
Procedure:
- Cell Isolation and Preparation: Isolate CD34+ HSCs from leukapheresis product using clinical-grade magnetic bead separation. Maintain cell viability >95% throughout processing.
- RNP Complex Formation: Combine high-purity Cas9 protein (100μM) with synthetic sgRNA (120μM) targeting the BCL11A enhancer sequence (5'-GCCCATTTACCCCTAGCTCC-3') in a 1:1.2 molar ratio. Incubate 10-20 minutes at room temperature.
- Electroporation: Resuspend 1×10^6 CD34+ cells in 100μL electroporation buffer. Add pre-formed RNP complex (at final concentration of 40μM Cas9) and transfer to electroporation cuvette. Execute pulse protocol (DT-130 for human CD34+ cells).
- Post-Edit Culture: Immediately transfer cells to pre-warmed culture medium supplemented with cytokines (100ng/mL SCF, 100ng/mL TPO, 100ng/mL FLT3-L). Culture at 37°C, 5% CO2 for 48 hours.
- Quality Control Assessments:
- Editing Efficiency: Assess by NGS of target locus (target: >80% indels)
- Viability: Measure by flow cytometry with viability dyes (target: >70% post-electroporation)
- Phenotypic Validation: Differentiate edited cells in erythroid conditions and measure HbF expression by HPLC (target: >25% HbF)
- Cryopreservation: Cryopreserve edited cells in controlled-rate freezer with DMSO for future infusion.
Critical Parameters: Maintain strict aseptic conditions throughout process; minimize time between cell collection and editing; use GMP-grade reagents; comprehensive viral safety testing.
Oncology Applications
Engineered Cell Therapies for Hematological Malignancies
CRISPR-Cas9 has revolutionized cancer immunotherapy through the development of enhanced chimeric antigen receptor (CAR) T-cell therapies. These approaches primarily focus on creating allogeneic ("off-the-shelf") CAR-T products by disrupting endogenous T-cell receptor (TCR) and human leukocyte antigen (HLA) genes to prevent graft-versus-host disease (GvHD) [78] [76]. Additionally, CRISPR editing can knockout inhibitory immune checkpoints (e.g., PD-1) to enhance antitumor activity and persistence of CAR-T cells [79].
Table 2: CRISPR-Engineered Cell Therapies in Oncology Clinical Trials
| Therapy | Target | Condition | Phase | Editing Strategy | Developer | Key Findings/Status |
|---|---|---|---|---|---|---|
| CTX112 | CD19 + multiple edits | B-cell malignancies, Autoimmune diseases | I/II | Allogeneic anti-CD19 CAR-T with immune evasion edits | CRISPR Therapeutics | RMAT designation; responses in patients refractory to T-cell engagers [52] |
| CTX131 | CD70 + multiple edits | Solid tumors, T-cell lymphomas | I/II | Allogeneic anti-CD70 CAR-T with enhanced potency edits | CRISPR Therapeutics | Updates expected in 2025 [52] |
| BEAM-201 | CD7 + multi-gene knockout | T-ALL, T-cell lymphoma | I/II | Anti-CD7 CAR-T with base editing (CD7, TRAC, CD52, PDCD1) | Beam Therapeutics | Reduced GvHD risk and improved allogeneic potential [79] |
| NK510 | Multiple NK cell targets | Advanced solid tumors | Preclinical (IND-cleared) | Base-edited natural killer cells | Base Therapeutics | Enhanced tumor recognition and killing [79] |
Experimental Protocol: Allogeneic CAR-T Cell Generation
Objective: Generate universal CAR-T cells through multiplexed gene editing to eliminate alloreactivity while maintaining antitumor efficacy.
Materials and Reagents:
- Source Cells: Healthy donor peripheral blood mononuclear cells (PBMCs)
- Editing Components: Cas9 protein, sgRNAs targeting TRAC, TRBC, B2M, PDCD1
- CAR Delivery: Lentiviral vector encoding anti-CD19 CAR
- Culture Media: TexMACS medium with IL-7/IL-15
- Activation Reagents: Clinical-grade anti-CD3/anti-CD28 beads
- Analytical Tools: Flow cytometry, cytotoxicity assays, cytokine release assays
Procedure:
- T-cell Isolation and Activation: Isolate CD3+ T-cells from PBMCs using magnetic separation. Activate with anti-CD3/anti-CD28 beads (bead:cell ratio 3:1) in TexMACS medium with 5% human AB serum and cytokines (10ng/mL IL-7, 10ng/mL IL-15).
- Multiplexed RNP Electroporation: At 48 hours post-activation, form RNP complexes with Cas9 and sgRNAs targeting TRAC (5'-GAGCAGGTCGCCACCATGAA-3'), B2M (5'-GTCTTTCAGCAAGGACTGGT-3'), and PDCD1 (5'-GACCTGCCGAGGCCACAGCC-3'). Electroporate using optimized T-cell protocol.
- CAR Transduction: At 24 hours post-electroporation, transduce cells with lentiviral vector encoding anti-CD19 CAR at MOI 5-10 in the presence of 8μg/mL polybrene. Centrifuge at 1000×g for 90 minutes (spinoculation).
- Expansion and Harvest: Culture cells in IL-7/IL-15 containing medium for 10-14 days, maintaining cell density at 0.5-2×10^6 cells/mL. Harvest when viability >90% and CAR expression >30%.
- Quality Control Assessments:
- Editing Efficiency: NGS of all target loci (target: >80% knockout for each gene)
- Alloreactivity Testing: Mixed lymphocyte reaction with donor PBMCs (target: >70% reduction in proliferation)
- Tumor Killing: Cytotoxicity assay against CD19+ tumor cells (target: >50% specific lysis at 10:1 E:T ratio)
- Cytokine Profile: Multiplex ELISA for IFN-γ, IL-2, IL-6 post-stimulation
- Sterility Testing: Mycoplasma, endotoxin, and sterility testing per FDA guidelines
Critical Parameters: Monitor for potential chromosomal abnormalities; validate absence of replication-competent lentivirus; functional potency assessment against relevant tumor models.
Metabolic Diseases
In Vivo Genome Editing for Metabolic Disorders
CRISPR-Cas9 applications for metabolic diseases increasingly employ in vivo editing approaches, particularly for liver-directed therapies. Lipid nanoparticle (LNP) delivery of CRISPR components enables transient but highly efficient editing of hepatocytes, offering potential one-time treatments for various metabolic disorders [9] [82]. This approach demonstrates particular promise for cardiovascular risk factors including familial hypercholesterolemia and elevated lipoprotein(a) [52] [83].
Table 3: In Vivo CRISPR Therapies for Metabolic and Cardiovascular Diseases
| Therapy | Target | Condition | Phase | Delivery | Developer | Key Findings/Status |
|---|---|---|---|---|---|---|
| VERVE-101 | PCSK9 | Heterozygous familial hypercholesterolemia | Ib (paused) | LNP with base editor | Verve Therapeutics | Enrollment paused due to laboratory abnormalities [83] |
| VERVE-102 | PCSK9 | HeFH, coronary artery disease | Ib | GalNAc-LNP with base editor | Verve Therapeutics | Well-tolerated in initial cohorts; update H1 2025 [83] |
| VERVE-201 | ANGPTL3 | Refractory hyperlipidemia, HoFH | Ib | GalNAc-LNP with base editor | Verve Therapeutics | First patient dosed November 2024 [83] |
| CTX310 | ANGPTL3 | Familial hypercholesterolemia, dyslipidemias | I | LNP with CRISPR-Cas9 | CRISPR Therapeutics | Preclinical NHP data showed durable protein reduction [52] [83] |
| CTX320 | LPA | Elevated lipoprotein(a) | I | LNP with CRISPR-Cas9 | CRISPR Therapeutics | Trial began 2024; update expected H1 2025 [52] |
| NTLA-2001 | TTR | Hereditary transthyretin amyloidosis | III | LNP with CRISPR-Cas9 | Intellia Therapeutics | ~90% TTR reduction sustained over 2 years; global Phase III ongoing [9] [83] |
Experimental Protocol: LNP Formulation for Liver-Directed Editing
Objective: Formulate and characterize LNPs encapsulating CRISPR-Cas9 mRNA and sgRNA for efficient in vivo hepatic gene editing.
Materials and Reagents:
- RNA Components: Cas9 mRNA (modified nucleotides, CleanCap), sgRNA (target-specific)
- Lipid Components: Ionizable lipid (e.g., DLin-MC3-DMA), phospholipid, cholesterol, PEG-lipid
- Formulation Equipment: Microfluidic mixer, TFF system, dynamic light scattering
- Analytical Tools: RiboGreen assay, HPLC, electron microscopy, animal models
Procedure:
- RNA Preparation: Prepare Cas9 mRNA with nucleotide modifications (e.g., pseudouridine, 5-methylcytidine) and CleanCap structure for enhanced stability and translation efficiency. Synthesize sgRNA with high-purity purification to minimize immune activation.
- LNP Formulation: Utilize microfluidic mixing technology with aqueous phase containing Cas9 mRNA and sgRNA at 0.2 mg/mL total RNA in citrate buffer (pH 4.0) and organic phase containing lipids in ethanol (ionizable lipid:phospholipid:cholesterol:PEG-lipid at 50:10:38.5:1.5 molar ratio). Use total flow rate of 12 mL/min with 3:1 aqueous:organic flow rate ratio.
- Buffer Exchange and Concentration: Dialyze formed LNPs against PBS (pH 7.4) using tangential flow filtration with 100 kDa molecular weight cutoff membrane. Concentrate to final RNA concentration of 1-2 mg/mL.
- LNP Characterization:
- Size and PDI: Dynamic light scattering (target: 70-90 nm, PDI <0.2)
- Encapsulation Efficiency: RiboGreen assay (target: >90%)
- RNA Integrity: Agarose gel electrophoresis or HPLC
- Morphology: Cryo-electron microscopy
- Endotoxin: LAL assay (target: <5 EU/mL)
- In Vivo Potency Testing: Administer LNP formulation to animal model (e.g., C57BL/6 mice) via intravenous injection at 1-3 mg/kg RNA dose. Assess editing efficiency in liver tissue after 7 days by NGS (target: >50% editing at therapeutic dose).
Critical Parameters: Maintain RNA integrity throughout process; ensure consistent LNP size distribution; comprehensive sterility testing; validate potency in relevant disease models.
The Scientist's Toolkit: Research Reagent Solutions
Table 4: Essential Reagents for CRISPR-Cas9 Clinical Translation
| Reagent/Category | Function | Key Considerations | Example Applications |
|---|---|---|---|
| GMP-grade Cas9 | RNA-guided DNA endonuclease | High purity, minimal endotoxin, comprehensive QC testing | Ex vivo cell therapies (CASGEVY) [79] |
| sgRNA/sgRNA | Target specificity and Cas9 recruitment | Chemical modifications for stability, HPLC purification | All CRISPR editing applications |
| Lipid Nanoparticles (LNPs) | In vivo delivery of CRISPR components | Liver tropism, encapsulation efficiency, safety profile | NTLA-2001, VERVE therapies [9] [82] |
| Electroporation Systems | Ex vivo delivery of RNP complexes | Cell viability optimization, clinical-scale capability | CAR-T engineering, HSC editing [76] |
| Base Editors | Chemical conversion of single nucleotides | Reduced indel formation, precision editing | VERVE-101, BEAM-101 [83] [79] |
| Prime Editors | Versatile precision editing without DSBs | Reverse transcriptase fusion, pegRNA design | PM359 for CGD [83] |
| AAV/Lentiviral Vectors | Delivery of editing components | Packaging capacity, immunogenicity, tropism | EBT-101 for HIV [79] |
| Analytical Tools | Quality control and safety assessment | NGS for on/off-target, cytogenetics, functional assays | Required for all IND applications |
CRISPR-Cas9 technology has unequivocally transitioned from basic research to clinical reality, with approved therapies demonstrating unprecedented efficacy for genetic diseases. The ongoing clinical trials across hematological, oncological, and metabolic disorders highlight the remarkable versatility of this platform. Current research focuses on enhancing precision through base and prime editing technologies, improving delivery efficiency with novel LNP formulations, and expanding the therapeutic landscape to include common complex diseases [9] [79].
Despite substantial progress, challenges remain in minimizing off-target effects, managing immune responses to editing components, and ensuring equitable access to these transformative therapies [78] [76]. The successful implementation of personalized in vivo CRISPR therapy for rare diseases, as demonstrated in the landmark case of an infant with CPS1 deficiency, further illustrates the potential for rapid development of bespoke genomic medicines [9]. As the field advances, continued innovation in delivery technologies and editing precision will undoubtedly expand the clinical applications of CRISPR-Cas9, ultimately fulfilling its potential to address previously untreatable human diseases.
The transformation of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) from a fundamental bacterial immune mechanism into a revolutionary genome-editing tool represents one of the most significant advancements in modern biotechnology [84]. This RNA-guided system provides researchers with unprecedented precision in modifying target genes, offering high accuracy and efficiency that has propelled the field from theoretical concept to clinical reality [84]. The core CRISPR-Cas9 system consists of two fundamental components: the Cas9 endonuclease, which creates double-strand breaks in DNA, and a guide RNA (gRNA) that directs the nuclease to specific genomic sequences through complementary base pairing [84].
The therapeutic potential of CRISPR-Cas9 lies in its ability to induce controlled DNA damage at precise genomic loci, harnessing the cell's endogenous repair mechanisms. The two primary repair pathways are non-homologous end joining (NHEJ), which often results in gene disruptions through small insertions or deletions, and homology-directed repair (HDR), which allows for precise gene corrections or insertions using a DNA template [84]. While NHEJ operates efficiently throughout the cell cycle, HDR is restricted primarily to late S and G2 phases, presenting particular challenges for editing non-dividing or slowly dividing cells [84].
The evolution of more precise editing technologies, including base editing and prime editing, has further expanded the therapeutic toolkit. Base editors utilize catalytically impaired Cas proteins fused to deaminase enzymes to directly convert one DNA base to another without creating double-strand breaks, while prime editors use Cas9 nickase fused to reverse transcriptase to enable more versatile genetic modifications [84]. These advancements have collectively driven the rapid clinical translation of CRISPR-based therapies, with the first approved treatments now available and an expanding pipeline of investigational therapies advancing through clinical development.
Approved CRISPR Therapies
Casgevy (exagamglogene autotemcel)
Casgevy represents the historic first FDA-approved therapy utilizing CRISPR-Cas9 technology, marking a watershed moment for the field [9] [85]. This ex vivo therapy is approved for the treatment of sickle cell disease (SCD) and transfusion-dependent beta thalassemia (TBT), representing a paradigm shift in the management of these inherited hemoglobinopathies [9].
The therapeutic mechanism involves the collection of a patient's own CD34+ hematopoietic stem cells, which are then genetically modified outside the body using CRISPR-Cas9. The editing process specifically targets the BCL11A gene, a key transcriptional regulator of fetal hemoglobin expression [85]. By disrupting an erythroid-specific enhancer in the BCL11A gene, the therapy reduces BCL11A expression, thereby reactivating the production of fetal hemoglobin, which does not exhibit the pathological polymerization characteristics of adult hemoglobin in SCD [85].
The manufacturing process involves electroporation of the CRISPR-Cas9 components into the harvested stem cells, followed by expansion and quality control verification before reinfusion into the patient. Notably, patients must undergo myeloablative conditioning with busulfan to clear bone marrow niche space for the engineered cells [9]. As of 2025, approximately 50 active clinical sites across North America, the European Union, and the Middle East are administering Casgevy, with significant progress made in securing reimbursement from state Medicaid programs in the United States and the United Kingdom's National Health Service [9].
Table 1: Currently Approved CRISPR-Based Therapies
| Therapy Name | Indication | Target | Mechanism | Approval Year |
|---|---|---|---|---|
| Casgevy (exa-cel) | Sickle cell disease, Transfusion-dependent beta thalassemia | BCL11A gene | Ex vivo disruption of BCL11A enhancer to increase fetal hemoglobin | 2023 [85] |
Late-Stage Clinical Candidates (Phase II/III)
The CRISPR therapeutic pipeline has expanded significantly beyond hematological disorders, with multiple candidates advancing into late-stage clinical trials across diverse disease areas including genetic disorders, cardiovascular diseases, and infectious diseases.
NTLA-2001 (Nexiguran Ziclumeran) for Transthyretin Amyloidosis
NTLA-2001, developed by Intellia Therapeutics in collaboration with Regeneron, represents a pioneering in vivo CRISPR-Cas9 therapy currently in Phase III trials for transthyretin amyloidosis (ATTR) [9] [83]. This systemic therapy utilizes lipid nanoparticles (LNPs) to deliver CRISPR-Cas9 components targeting the TTR gene in hepatocytes, the primary source of transthyretin protein production [9].
The MAGNITUDE Phase III clinical trial is evaluating a single dose of NTLA-2001 compared to placebo in more than 700 patients with either hereditary (ATTRv) or wild-type (ATTRwt) forms of the disease [83]. Previous Phase I results demonstrated rapid, deep, and durable reductions in TTR protein levels, with participants showing an average of approximately 90% reduction in serum TTR concentration that remained sustained throughout the trial duration [9]. All 27 participants who reached two years of follow-up maintained this response without evidence of diminishing effect [9]. Functional and quality-of-life assessments have largely shown disease stabilization or improvement in symptoms, with a safety profile characterized primarily by mild to moderate infusion-related events [9].
NTLA-2002 (Lonvoguran Ziclumeran) for Hereditary Angioedema
NTLA-2002 is another in vivo LNP-delivered CRISPR therapy from Intellia Therapeutics that has advanced to Phase III trials for hereditary angioedema (HAE) [9] [85] [86]. This therapy targets the kallikrein B1 (KLKB1) gene, which encodes the precursor of plasma kallikrein, a key mediator of the inflammatory pathways responsible for HAE attacks [9] [83].
Phase I/II trial results published in the New England Journal of Medicine demonstrated that participants receiving the higher dose achieved an average 86% reduction in kallikrein levels and a significant decrease in the frequency of HAE attacks [9]. Eight of eleven participants in the high-dose group remained completely attack-free during the 16-week observation period reported [9]. The therapy has received multiple regulatory designations including Orphan Drug, RMAT, and PRIME status, potentially positioning it to become the first one-time treatment for HAE, with a possible U.S. launch by 2027 [86]. Intellia completed enrollment for the global Phase III HAELO study in September 2025 [85].
LBP-EC01 for Urinary Tract Infections
LBP-EC01, developed by Locus Biosciences, represents a novel application of CRISPR technology targeting bacterial infections [85]. This therapy utilizes CRISPR-Cas3-enhanced bacteriophages specifically engineered to target and eliminate antimicrobial-resistant Escherichia coli causing uncomplicated urinary tract infections (uUTIs) [85].
Unlike traditional CRISPR-Cas9 systems that create precise double-strand breaks, the Cas3 enzyme creates larger deletions in bacterial DNA, resulting in potent antibacterial activity [85]. Positive results from Part 1 of the Phase II ELIMINATE trial demonstrated significant reduction in bacterial load and clinical improvement in patients with multidrug-resistant E. coli infections [85]. This approach potentially offers a solution to the growing challenge of antimicrobial resistance by providing a targeted therapeutic that can be tailored to specific bacterial pathogens while sparing beneficial microbiota.
Table 2: Late-Stage CRISPR Clinical Candidates (Phase II/III)
| Therapy Name | Developer | Indication | Target | Delivery Method | Trial Phase |
|---|---|---|---|---|---|
| NTLA-2001 | Intellia Therapeutics | Transthyretin Amyloidosis (ATTR) | TTR gene | LNP (in vivo) | Phase III [83] |
| NTLA-2002 | Intellia Therapeutics | Hereditary Angioedema (HAE) | KLKB1 gene | LNP (in vivo) | Phase III [9] [85] |
| LBP-EC01 | Locus Biosciences | Urinary Tract Infections (E. coli) | Bacterial DNA | Bacteriophage (in vivo) | Phase II/III [85] |
| CB-010 | Caribou Biosciences | Systemic Lupus Erythematosus | CD19+ B-cells | Ex vivo CAR-T | Phase I [85] |
Emerging Technologies and Delivery Systems
Advancements in Delivery Platforms
The successful clinical translation of CRISPR therapies depends critically on efficient delivery systems that can safely and precisely deliver editing components to target cells [86]. While ex vivo approaches like those used in Casgevy involve editing cells outside the body before reinfusion, in vivo therapies require sophisticated delivery vehicles to protect the editing machinery and facilitate cellular uptake [86].
Lipid nanoparticles (LNPs) have emerged as a leading delivery platform for in vivo CRISPR therapies, particularly for liver-targeted applications [9] [86]. These nanocarriers form protective vesicles around CRISPR components and naturally accumulate in hepatocytes following systemic administration [9]. Recent advancements include the development of novel biodegradable ionizable lipids that improve safety profiles and enhance delivery efficiency. Researchers at the University of Toronto have identified an LNP-formulated ionizable lipid (A4B4-S3) that outperforms the clinical benchmark lipid (SM-102) used in Moderna's COVID-19 vaccine for hepatic mRNA delivery in murine models [86].
Viral vectors, particularly adeno-associated viruses (AAVs), continue to play an important role in delivery, especially for tissues beyond the liver. However, immune responses to viral components and potential integration events remain concerns that limit redosing possibilities [9]. LNPs offer advantages in this regard, as demonstrated by the successful redosing of participants in Intellia's hATTR trial and the personalized CPS1 deficiency treatment, where multiple administrations were safely delivered to enhance editing efficiency [9].
Tissue-Specific Targeting and Novel Nuclease Platforms
Emerging technologies are expanding the possibilities for tissue-specific CRISPR delivery beyond the liver. UK researchers have developed CRISPR MiRAGE (miRNA-activated genome editing), a system that leverages tissue-specific microRNA signatures to restrict editing activity to particular cell types [86]. This approach has been successfully demonstrated in mouse models of Duchenne muscular dystrophy, showing enhanced specificity and reduced off-target effects [86].
The CRISPR toolkit has also expanded beyond standard Cas9 nucleases. Base editing platforms, which enable precise single-nucleotide conversions without creating double-strand breaks, are advancing through clinical trials [84]. Verve Therapeutics' VERVE-101, an adenine base editor targeting the PCSK9 gene for familial hypercholesterolemia, represents the first base editing approach to enter clinical testing, though enrollment was temporarily paused due to laboratory abnormalities [83]. Prime editing systems offer even greater precision and versatility, capable of making all possible nucleotide substitutions as well as small insertions and deletions without double-strand break formation [84].
Experimental Protocols and Methodologies
LNP-Mediated In Vivo CRISPR Delivery Protocol
The success of liver-directed CRISPR therapies like NTLA-2001 and NTLA-2002 relies on optimized LNP delivery protocols. The following methodology outlines the standard procedure for LNP formulation and in vivo administration based on current clinical approaches:
Guide RNA Design and Validation: Design single guide RNAs (sgRNAs) with full complementarity to the target genomic sequence (typically 20 nucleotides) followed by an appropriate protospacer adjacent motif (PAM) sequence. Validate sgRNA specificity using computational prediction tools (e.g., CrisprScan, ChopChop) and empirically test editing efficiency in relevant cell lines using targeted deep sequencing.
mRNA and gRNA Preparation: In vitro transcribe and purify Cas9 mRNA with complete 5' capping and 3' polyadenylation. Synthesize and purify sgRNA using T7 polymerase-based transcription. Ensure pharmaceutical-grade purity and endotoxin testing for clinical applications.
LNP Formulation: Prepare lipid mixtures containing ionizable cationic lipid, phospholipid, cholesterol, and PEG-lipid in ethanol phase at molar ratios optimized for hepatic delivery. Combine the lipid mixture with aqueous phase containing Cas9 mRNA and sgRNA at specific ratios using microfluidic mixing devices. Typical N:P ratios (nitrogen atoms in cationic lipid to phosphate atoms in RNA) range from 3:1 to 6:1.
LNP Characterization and Quality Control: Determine particle size and polydispersity index using dynamic light scattering (target size: 70-100 nm). Measure zeta potential and encapsulation efficiency. Validate sterility, endotoxin levels, and RNA integrity before in vivo administration.
In Vivo Administration: Administer LNP formulations via intravenous injection at dosages typically ranging from 0.1 to 1.0 mg RNA per kg body weight. Monitor animals or patients for potential infusion-related reactions and assess editing efficiency at predetermined timepoints through tissue biopsy or blood-based biomarkers.
Analysis of Editing Efficiency and Specificity
Robust assessment of CRISPR editing outcomes is essential for therapeutic development:
Targeted Deep Sequencing: Amplify target genomic regions using PCR primers flanking the edit site. Incorporate unique molecular identifiers (UMIs) to reduce amplification bias and enable precise quantification of editing efficiencies. Sequence libraries using Illumina platforms and analyze sequences for insertion/deletion patterns (for nuclease approaches) or base conversion rates (for base editing approaches).
Off-Target Analysis: Identify potential off-target sites using computational prediction tools (e.g., Cas-OFFinder) and in vitro cleavage assays (GUIDE-seq, CIRCLE-seq). Validate potential off-target sites in treated cells or tissues using targeted amplicon sequencing.
Functional Assessment: Quantify target protein reduction (e.g., TTR for NTLA-2001, kallikrein for NTLA-2002) using ELISA or mass spectrometry. For ex vivo therapies, assess functional correction through appropriate cellular assays (e.g., hemoglobin profiling for sickle cell therapies).
Table 3: Essential Research Reagents for CRISPR Clinical Development
| Reagent Category | Specific Examples | Function | Clinical Relevance |
|---|---|---|---|
| Nuclease Systems | Cas9 mRNA, Base editor proteins | Catalyze DNA cleavage or modification | Core therapeutic component |
| Guide RNAs | Chemically modified sgRNAs | Target specificity through complementary binding | Determines genomic target |
| Delivery Vehicles | Ionizable lipids, AAV capsids | Protect and deliver editing components | Enables in vivo administration |
| Analytical Tools | NGS assays for on/off-target, ELISA for protein quantification | Assess editing efficiency and safety | Critical for potency and safety evaluation |
| Cell Culture Systems | Primary hepatocytes, CD34+ HSPCs | Model systems for optimization | Ex vivo therapy development |
The CRISPR clinical trial landscape in 2025 reflects a maturing field that has successfully transitioned from proof-of-concept studies to approved therapies and a diverse late-stage pipeline. The landmark approval of Casgevy for sickle cell disease and beta thalassemia established CRISPR as a validated therapeutic platform, while the rapid advancement of in vivo approaches like NTLA-2001 and NTLA-2002 demonstrates the field's capacity to address increasingly complex therapeutic challenges.
Several key trends are shaping the future of CRISPR therapeutics. First, the expansion beyond monogenic diseases to common conditions like cardiovascular disease (e.g., Verve's programs targeting PCSK9 and ANGPTL3) and infectious diseases (e.g., Locus's CRISPR-enhanced bacteriophages) significantly broadens the potential patient populations that could benefit from CRISPR interventions [87] [85]. Second, continued innovation in delivery technologies, including tissue-specific LNPs and novel viral vectors, is gradually overcoming the historic challenge of targeted delivery. Third, the emergence of more precise editing platforms like base and prime editors offers the potential for enhanced safety profiles and expanded therapeutic applications.
However, significant challenges remain. Delivery efficiency to non-hepatic tissues, potential immunogenicity of bacterial-derived Cas proteins, and the long-term persistence of editing effects require continued investigation [84]. The field must also address manufacturing scalability and cost considerations to ensure equitable access to these potentially transformative therapies. The recent reductions in biotechnology venture capital funding and government science budgets noted in 2025 could potentially impact the pace of future innovation [9].
Despite these challenges, the remarkable progress in CRISPR-based therapeutics over the past decade provides strong rationale for optimism. As the field continues to mature, the convergence of improved editing precision, advanced delivery systems, and enhanced safety profiles positions CRISPR therapies to potentially address some of medicine's most intractable genetic disorders, fundamentally transforming the treatment paradigm for patients worldwide.
Overcoming CRISPR Challenges: Off-Target Effects, Delivery Hurdles, and Specificity Enhancement
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) system has revolutionized genome engineering, providing an unprecedented tool for precise gene editing in biomedical research and therapeutic development. This bacterial adaptive immune system functions as a ribonucleoprotein complex where the Cas9 nuclease is directed by a single-guide RNA (sgRNA) to create double-strand breaks (DSBs) at specific genomic loci complementary to the sgRNA sequence and adjacent to a Protospacer Adjacent Motif (PAM) [88] [89]. The simplicity, efficiency, and programmability of CRISPR-Cas9 have made it the preferred genome editing technology, surpassing earlier tools like Zinc Finger Nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs) [89].
Despite its transformative potential, a significant challenge remains: off-target effects [90] [88]. These occur when the Cas9 nuclease cleaves DNA at unintended genomic sites with sequences similar to the target site, leading to unintended mutations that can compromise experimental integrity and pose serious safety risks in therapeutic applications [88] [91]. Off-target effects primarily arise from the system's tolerance for mismatches between the sgRNA and target DNA, particularly when mismatches occur in the distal region from the PAM sequence, with Cas9 capable of tolerating up to 3-6 base pair mismatches [88] [92]. Additional contributing factors include non-canonical PAM recognition (e.g., 'NAG' or 'NGA' instead of 'NGG'), DNA/RNA bulges from imperfect complementarity, and genetic variations such as single nucleotide polymorphisms (SNPs) that can create novel off-target sites [91] [92].
The clinical significance of off-target effects was highlighted during the regulatory review of Casgevy (exa-cel), the first FDA-approved CRISPR-based therapy, where comprehensive off-target characterization was required [91]. Unintended edits in protein-coding regions, particularly in oncogenes or tumor suppressor genes, could potentially initiate tumorigenesis, while large structural variations including chromosomal translocations and megabase-scale deletions present additional safety concerns [27]. Consequently, rigorous detection and mitigation of off-target effects are essential components of CRISPR experimental design and therapeutic development.
Mechanisms and Types of Off-Target Effects
Molecular Basis of Off-Target Activity
The precision of CRISPR-Cas9 is governed by the seed sequence - the PAM-proximal 10-12 nucleotide region of the sgRNA that is crucial for specific target recognition and cleavage [92]. While perfect complementarity in this region typically ensures efficient on-target cleavage, mismatches in regions distal to the PAM are more tolerated, leading to potential off-target activity [92]. The structural biology of Cas9 reveals a conformational activation mechanism wherein complementarity in the seed region triggers structural rearrangements that activate the nuclease domains, explaining why mismatches in this region are less tolerated [88].
The PAM recognition mechanism serves as the initial gatekeeper for Cas9 activity. While SpCas9 primarily recognizes the canonical 'NGG' PAM, it can also tolerate non-canonical variants like 'NAG' and 'NGA' with reduced efficiency, substantially expanding the potential off-target landscape within the genome [92]. Emerging "PAM-relaxed" or "PAM-free" Cas variants, such as SpRY and SpCas9-NG, further expand targeting range but may increase off-target potential without proper optimization [92].
Categories of Genomic Alterations
- Simple Indels: Small insertions or deletions represent the most common outcome of error-prone non-homologous end joining (NHEJ) repair at both on-target and off-target sites. These are typically detected by amplicon sequencing but may be overestimated when larger deletions go undetected [27].
- Large Structural Variations (SVs): Recent studies reveal that CRISPR editing can induce kilobase- to megabase-scale deletions, chromosomal losses, and truncations that often escape conventional detection methods [27]. These substantial rearrangements result from multiple DSBs and aberrant DNA repair processes.
- Chromosomal Translocations: Simultaneous cleavage at on-target and off-target sites can lead to potentially oncogenic translocations between heterologous chromosomes [27]. Methods like CAST-Seq and LAM-HTGTS were specifically developed to detect these rearrangements [27].
- Complex Genomic Rearrangements: Phenomena like chromothripsis (massive chromosomal shattering and reorganization) have been observed in CRISPR-edited cells, representing worst-case scenarios for genomic instability [27].
Table 1: Categories of Unintended Genomic Alterations in CRISPR Editing
| Category | Scale | Detection Methods | Potential Functional Impact |
|---|---|---|---|
| Point mutations | Single bases | WGS, targeted sequencing | Altered protein function, regulatory changes |
| Small indels | 1-100 bp | Amplicon sequencing, ICE analysis | Frameshifts, gene disruption |
| Large deletions | >100 bp | Long-read sequencing, CAST-Seq | Gene loss, regulatory region deletion |
| Chromosomal translocations | Inter-chromosomal | CAST-Seq, LAM-HTGTS | Oncogenic fusions, genomic instability |
| Complex rearrangements | Multiple loci | WGS, structural variation analysis | Chromothripsis, cellular transformation |
Computational Prediction Methods
Algorithm-Based Prediction Tools
Computational prediction represents the first line of defense against off-target effects, employing algorithms to identify potential off-target sites based on sequence similarity to the intended target [88] [92]. These tools can be categorized into two primary classes:
Alignment-based models perform exhaustive searches through reference genomes to identify sites with partial complementarity to the sgRNA. Key tools in this category include:
- CasOT: The first exhaustive tool for off-target prediction that allows custom adjustment of parameters including PAM sequence and mismatch number (up to 6 mismatches) [88].
- Cas-OFFinder: A widely applied tool with high tolerance for sgRNA length, PAM types, and the number of mismatches or bulges [88].
- FlashFry: A high-throughput tool capable of characterizing hundreds of thousands of CRISPR target sequences rapidly while providing GC content information and on/off-target scores [88].
- Crisflash: Distinguished by computational efficiency, being over an order of magnitude faster than other software while providing both sgRNA design and latent off-target discovery [88].
Scoring-based models employ more sophisticated algorithms that weight various factors to prioritize potential off-target sites:
- MIT Scoring Algorithm: Incorporates the position-specific effect of mismatches relative to the PAM, with mismatches distal to the PAM being more tolerated [88].
- CCTop (Consensus Constrained TOPology prediction): Utilizes distances of mismatches from the PAM in its prediction model [88].
- CFD (Cutting Frequency Determination): Based on experimentally validated datasets to improve prediction accuracy [88].
- DeepCRISPR: Incorporates both sequence and epigenetic features using deep learning to enhance prediction capability [88].
Emerging Deep Learning Approaches
Recent advances in artificial intelligence have yielded more sophisticated prediction tools such as CCLMoff, which leverages deep learning and RNA language models to predict off-target effects with improved accuracy across diverse datasets [93]. This approach addresses a critical limitation of traditional methods that perform poorly on previously unseen guide RNA sequences. By incorporating a pre-trained RNA language model from RNAcentral, CCLMoff captures complex sequence relationships between guide RNAs and potential target sites, demonstrating superior generalization capabilities across multiple next-generation sequencing-based detection datasets [93].
Computational Prediction Workflow for CRISPR Off-Target Effects
Limitations of Computational Prediction
While in silico tools provide valuable initial guidance, they possess significant limitations. Most algorithms are biased toward sgRNA-dependent off-target effects and insufficiently consider the complex intranuclear microenvironment, including epigenetic states and chromatin organization [88]. Additionally, their performance depends heavily on the quality and completeness of reference genomes, and they may fail to account for individual genetic variations that could create novel off-target sites [92]. Therefore, computational predictions must be complemented with experimental validation, particularly for therapeutic applications.
Experimental Detection Methods
Cell-Free Detection Methods
Cell-free methods utilize purified genomic DNA or cell-free chromatin incubated with Cas9-sgRNA ribonucleoproteins (RNPs) to identify potential cleavage sites without cellular constraints.
- Digenome-seq: This was the first in vitro off-target assay developed, involving in vitro digestion of purified genomic DNA using Cas9/sgRNA complexes followed by whole-genome sequencing to detect cleavage sites [88] [92]. While highly sensitive, it requires high sequencing coverage and a reference genome, and does not account for chromatin accessibility [88].
- CIRCLE-seq: An enhanced method that circularizes sheared genomic DNA before incubation with Cas9/sgRNA RNP, then linearizes cleaved DNA for sequencing [88] [91]. This approach offers high sensitivity and reduced background, capable of detecting low-frequency off-target sites.
- SITE-seq: A biochemical method employing selective biotinylation and enrichment of fragments after Cas9/gRNA digestion, requiring minimal read depth and no reference genome but suffering from lower sensitivity and validation rate [88].
- DIG-seq: An adaptation of Digenome-seq that uses cell-free chromatin to account for chromatin accessibility, resulting in higher validation rates than the original method [88].
Cell Culture-Based Methods
These methods detect off-target effects within living cells, capturing the complexity of cellular environments including chromatin organization and DNA repair mechanisms.
- GUIDE-seq (Genome-wide Unbiased Identification of DSBs Enabled by Sequencing): Utilizes integration of double-stranded oligodeoxynucleotides (dsODNs) into DSBs followed by enrichment and sequencing [88] [91]. This method is highly sensitive, cost-effective, and has low false positive rates, though it is limited by transfection efficiency [88].
- BLESS (Direct In Situ Breaks Labeling, Enrichment on Streptavidin and Next-Generation Sequencing): Captures DSBs in situ using biotinylated adaptors in fixed cells, allowing real-time detection of breaks but only identifying off-target sites present at the time of detection [88] [92].
- BLISS (Break Labeling In Situ and Sequencing): Similar to BLESS but uses dsODNs with T7 promoter sequences to capture DSBs in situ, requiring low input material [88].
- Discover-seq: Utilizes the DNA repair protein MRE11 as bait to perform ChIP-seq, offering high sensitivity and precision in cells but with some false positives [88].
- LAM-HTGTS: Detects DSB-caused chromosomal translocations by sequencing bait-prey DSB junctions, accurately identifying chromosomal rearrangements but limited to DSBs with translocation events [88].
In Vivo Detection Methods
As CRISPR therapies advance, detecting off-target effects in living organisms has become increasingly important.
- GUIDE-tag: An adaptation of GUIDE-seq for in vivo applications that uses biotin-dsDNA to mark DSBs, offering high sensitivity for detecting off-target sites in vivo though with relatively low incorporation rate (~6%) [88].
- VIVO (Verification of In Vivo Off-targets): Combines CIRCLE-seq with in vivo delivery to identify potential off-target sites that may be relevant in therapeutic contexts.
Table 2: Experimental Methods for Detecting CRISPR Off-Target Effects
| Method | Type | Sensitivity | Advantages | Limitations |
|---|---|---|---|---|
| Digenome-seq | Cell-free | High | Genome-wide, sensitive | Expensive, high coverage needed, no chromatin context |
| CIRCLE-seq | Cell-free | Very high | Low background, high sensitivity | In vitro system, may overpredict sites |
| GUIDE-seq | Cell-based | High | Low false positives, cost-effective | Limited by transfection efficiency |
| BLESS/BLISS | Cell-based | Medium | Captures DSBs in situ | Snapshots only, may miss transient events |
| Discover-seq | Cell-based | High | Uses endogenous repair machinery | Some false positives |
| CAST-Seq | Cell-based | High for SVs | Detects chromosomal rearrangements | Focused on structural variations |
| Whole Genome Sequencing | Comprehensive | Ultimate | Detects all mutation types | Very expensive, data analysis challenges |
Experimental Methods for Detecting Off-Target Effects
Risk Assessment and Mitigation Strategies
Comprehensive Risk Assessment Framework
A robust risk assessment strategy for CRISPR applications requires a multi-layered approach that considers both the specific application and the potential consequences of off-target effects.
Application-Based Risk Considerations:
- Basic Research: For functional genomics studies where off-target effects could confound phenotypic interpretations, candidate site sequencing based on computational predictions may be sufficient [91].
- Ex Vivo Therapeutics: In clinical applications like CAR-T cell engineering or stem cell therapies where editing occurs outside the body, intermediate scrutiny using methods like GUIDE-seq or amplicon sequencing of predicted off-target sites is appropriate, coupled with single-cell cloning to select correctly edited cells [91].
- In Vivo Therapeutics: For direct in vivo gene editing where selection of correctly edited cells is impossible, the most rigorous assessment using multiple orthogonal methods (e.g., CIRCLE-seq combined with in vivo validation) is essential [91] [27].
Biological Context Risk Factors:
- Target Gene Function: Editing tumor suppressor genes or oncogenes requires heightened scrutiny due to the potentially severe consequences of off-target activity [27].
- Cell Type and Proliferation Status: Actively dividing cells may dilute or fix off-target mutations differently than non-dividing cells, affecting risk calculations [27].
- Therapeutic Window: The balance between therapeutic benefit and potential risk determines the acceptable off-target rate [27].
Strategies for Minimizing Off-Target Effects
Several effective strategies have been developed to reduce the likelihood of off-target editing:
CRISPR System Selection and Engineering:
- High-Fidelity Cas Variants: Engineered Cas9 variants like SpCas9-HF1, eSpCas9, and HiFi Cas9 demonstrate reduced off-target activity while maintaining on-target efficiency [91] [92].
- Alternative Cas Nucleases: Cas12a (Cpf1) and other Cas proteins with different PAM requirements or increased specificity can reduce off-target potential [88] [91].
- DNA Nickase Approaches: Using Cas9 nickases (nCas9) that create single-strand breaks instead of DSBs, either as standalone systems or in paired configurations, significantly reduces off-target effects [91] [92].
- Base and Prime Editing: These more recent technologies enable precise nucleotide changes without creating DSBs, substantially reducing the risk of off-target effects and structural variations [91] [27].
Guide RNA Optimization:
- Truncated sgRNAs: Shorter guide RNAs (17-18 nucleotides instead of 20) demonstrate reduced off-target activity while often maintaining on-target efficiency [92].
- Chemical Modifications: Incorporating 2'-O-methyl analogs (2'-O-Me) and 3' phosphorothioate bonds (PS) in synthetic gRNAs can reduce off-target edits and increase on-target efficiency [91].
- GC Content Optimization: Guides with higher GC content in the seed region stabilize the DNA:RNA duplex, increasing specificity [91].
Delivery and Expression Optimization:
- RNP Delivery: Using preassembled Cas9-gRNA ribonucleoprotein complexes rather than plasmid DNA encoding CRISPR components reduces persistence time in cells, limiting off-target opportunities [91].
- Regulatable Systems: Inducible or self-inactivating CRISPR systems that limit the temporal window of editing activity can minimize off-target effects [91].
- Dosage Optimization: Using the minimum effective concentration of CRISPR components reduces off-target editing while maintaining on-target efficiency [91].
The Scientist's Toolkit: Essential Reagents and Methods
Table 3: Research Reagent Solutions for Off-Target Assessment
| Reagent/Method | Function | Application Context |
|---|---|---|
| High-Fidelity Cas9 Variants (e.g., SpCas9-HF1, HiFi Cas9) | Engineered nucleases with reduced off-target activity | All applications requiring enhanced specificity |
| Chemically Modified sgRNAs (2'-O-Me, PS bonds) | Enhanced stability and specificity | Therapeutic applications, sensitive cell types |
| Cas9 Nickase (nCas9) | Creates single-strand breaks instead of DSBs | Applications where paired nicking is feasible |
| Ribonucleoprotein (RNP) Complexes | Precomplexed Cas9 and sgRNA for transient expression | Reduced off-target effects, primary cell editing |
| GUIDE-seq Oligonucleotides | Double-stranded oligos for DSB tagging | Comprehensive off-target mapping in cells |
| CIRCLE-seq Kits | In vitro off-target detection kits | Preclinical safety assessment |
| CAST-Seq Reagents | Detection of chromosomal rearrangements | Assessment of structural variations |
| ICE Analysis Tool (Inference of CRISPR Edits) | Computational analysis of editing efficiency | Rapid assessment of editing outcomes from Sanger sequencing |
The remarkable precision of CRISPR-Cas9 genome editing has revolutionized biomedical research and therapeutic development, yet off-target effects remain a significant consideration for rigorous experimental design and clinical translation. A comprehensive approach to understanding and measuring these effects integrates computational prediction with experimental validation, selecting methods appropriate for the specific application and risk threshold. While significant progress has been made in developing sensitive detection technologies and high-fidelity editing systems, the field continues to evolve with emerging challenges such as large structural variations and complex genomic rearrangements receiving increased attention.
The future of CRISPR safety assessment lies in the development of more predictive algorithms incorporating epigenetic and structural genomic features, more sensitive detection methods capable of identifying rare off-target events in heterogeneous cell populations, and continued innovation in editor engineering to maximize specificity without compromising efficiency. As the CRISPR landscape expands to include newer technologies like base editing, prime editing, and epigenome editing, each platform will require tailored off-target assessment strategies. Through rigorous characterization and mitigation of off-target effects, researchers can fully harness the transformative potential of CRISPR technologies while ensuring their safe application in both basic research and clinical therapeutics.
The CRISPR-Cas9 system has emerged as a revolutionary genome-editing tool with profound implications for basic research and therapeutic development. At its core, the system consists of a Cas9 nuclease and a guide RNA (gRNA) that directs the nuclease to a specific DNA sequence complementary to the gRNA, adjacent to a Protospacer Adjacent Motif (PAM) [94] [44]. While the wild-type Streptococcus pyogenes Cas9 (SpCas9) serves as a powerful workhorse, its application is constrained by a significant limitation: off-target effects. These occur when the Cas9-gRNA complex binds and cleaves DNA at sites with sequence similarity to the intended target, leading to unintended mutations that can confound experimental results and pose serious safety risks in clinical applications [94] [95].
To address this critical issue, protein engineering strategies have been employed to develop high-fidelity Cas9 variants with dramatically improved specificity. Among these, eSpCas9, SpCas9-HF1, and HypaCas9 represent landmark achievements. These engineered variants are designed to minimize non-specific interactions with DNA while maintaining robust on-target activity, thereby expanding the potential for safe and effective therapeutic genome editing [96] [95] [97]. This guide provides an in-depth technical examination of these three variants, detailing their engineering rationales, molecular mechanisms, and experimental validation for researchers and drug development professionals.
Engineering Strategies and Molecular Mechanisms
The development of high-fidelity Cas9 variants has relied primarily on structure-guided rational design to reduce the enzyme's promiscuity while preserving its catalytic efficiency.
SpCas9-HF1: Attenuating Non-Specific DNA Contacts
SpCas9-HF1 (High-Fidelity 1) was engineered based on the "excess energy" hypothesis, which posits that the wild-type SpCas9-sgRNA complex possesses more binding energy than is strictly necessary for on-target cleavage, enabling it to tolerate mismatches at off-target sites [96]. To reduce this excess energy, the developers targeted residues involved in non-specific interactions with the DNA phosphate backbone.
- Engineering Rationale: Structural studies identified four key residues (N497, R661, Q695, and Q926) that form hydrogen bonds with the target DNA strand. Alanine substitutions (N497A, R661A, Q695A, Q926A) were designed to disrupt these non-essential contacts, thereby increasing the stringency for target recognition [96] [95].
- Mechanistic Insight: Single-molecule FRET (smFRET) studies later revealed that SpCas9-HF1 does not reduce DNA binding affinity but instead raises the activation threshold for the catalytic HNH nuclease domain. When bound to mismatched off-target DNA, the HNH domain of SpCas9-HF1 is trapped in an inactive conformational state, preventing cleavage [97].
eSpCas9: Weakening gRNA-DNA Heteroduplex Stability
eSpCas9 (enhanced Specificity Cas9) was developed concurrently and operates on a similar principle of reducing non-productive DNA interactions.
- Engineering Rationale: The design focused on mutating positively charged residues (K810A/K1003A/R1060A in version 1.0; K848A/K1003A/R1060A in version 1.1) that interact with the DNA non-target strand. These mutations were predicted to weaken the stability of the unwound DNA heteroduplex, particularly at off-target sites where base pairing is imperfect [95].
- Mechanistic Insight: Like SpCas9-HF1, eSpCas9(1.1) employs a conformational proofreading mechanism. Its REC3 domain exhibits altered dynamics, which reduces the propensity of the HNH domain to adopt the active conformation on mismatched substrates [97] [98].
HypaCas9: Exploiting Allosteric Regulation of HNH
HypaCas9 (Hyper-accurate Cas9) was designed through a systematic exploration of residues involved in allosteric regulation of nuclease activity.
- Engineering Rationale: Building on insights from SpCas9-HF1 and eSpCas9, researchers identified a cluster of residues in the REC3 domain (N692, M694, Q695, H698) critical for sensing DNA complementarity. Alanine substitutions at these positions (N692A/M694A/Q695A/H698A) enhance the enzyme's ability to discriminate against mismatched targets by modulating the conformational pathway leading to HNH activation [95] [97].
- Mechanistic Insight: The REC3 domain acts as an allosteric effector that recognizes the RNA-DNA heteroduplex and governs HNH activation. The HypaCas9 mutations tighten this allosteric control, requiring more perfect complementarity to trigger the conformational change that activates cleavage [97].
Table 1: Engineering Strategies and Key Mutations of High-Fidelity Cas9 Variants
| Variant | Year | Engineering Strategy | Key Mutations | Structural Domains Affected |
|---|---|---|---|---|
| SpCas9-HF1 | 2016 | Structure-guided disruption of DNA backbone contacts | N497A, R661A, Q695A, Q926A | REC2, REC3, HNH-RuvC Linker |
| eSpCas9(1.1) | 2016 | Weakening non-target strand interactions | K848A, K1003A, R1060A | REC3, RuvC |
| HypaCas9 | 2017 | Allosteric control of HNH via REC3 domain | N692A, M694A, Q695A, H698A | REC3 |
The following diagram illustrates the shared conformational proofreading mechanism by which these high-fidelity variants achieve their specificity, by imposing a stricter barrier to HNH nuclease activation.
Experimental Validation and Performance Metrics
Rigorous biochemical and cellular assays have been employed to quantify the performance of these variants, confirming their high fidelity without substantial compromise of on-target activity.
Biochemical Characterization
In vitro cleavage assays using purified proteins demonstrate that all three variants cleave on-target DNA with kinetics comparable to wild-type SpCas9. However, their activities on mismatched DNA substrates are significantly reduced. For instance, SpCas9-HF1 exhibited cleavage rates on off-target substrates that were often undetectable, while maintaining >85% of wild-type on-target efficiency for the majority of gRNAs tested [96].
Cellular On-Target and Off-Target Assessment
Validation in human cells is crucial for assessing real-world performance. The GUIDE-seq (Genome-wide, Unbiased Identification of DSBs Enabled by sequencing) method has been widely used for this purpose.
- SpCas9-HF1 Performance: In one seminal study, GUIDE-seq was performed with eight different sgRNAs. Wild-type SpCas9 induced 2 to 25 off-target sites per sgRNA. In stark contrast, SpCas9-HF1 reduced off-target effects to undetectable levels for six of the seven sgRNAs that showed off-target activity with wild-type, and only a single off-target site was detected for the remaining sgRNA [96].
- HypaCas9 and eSpCas9 Performance: Both variants similarly demonstrate a dramatic reduction in genome-wide off-target events across multiple endogenous gene targets. HypaCas9, for example, was shown to retain high on-target activity (>70% of wild-type) at the majority of sites tested while exhibiting exceptional specificity [97].
Table 2: Performance Comparison of High-Fidelity Cas9 Variants in Human Cells
| Variant | On-Target Efficiency (Relative to WT) | Off-Target Reduction | Key Validation Assays |
|---|---|---|---|
| SpCas9-HF1 | >70% for 32/37 sgRNAs tested [96] | Undetectable for 6/7 sgRNAs with WT off-targets [96] | GUIDE-seq, T7EI, deep sequencing |
| eSpCas9(1.1) | >70% for 23/24 endogenous sites [97] | Significant reduction, profile similar to SpCas9-HF1 [97] | GUIDE-seq, T7EI, deep sequencing |
| HypaCas9 | >70% for 19/24 endogenous sites [97] | Superior to SpCas9-HF1 for some mismatches [97] | smFRET, EGFP disruption, GUIDE-seq |
Detailed Experimental Protocol: Evaluating Variant Specificity Using GUIDE-seq
The following protocol provides a methodology for assessing the genome-wide specificity of high-fidelity Cas9 variants in human cells, adapted from studies that validated eSpCas9, SpCas9-HF1, and HypaCas9 [96] [97].
Materials and Reagents
- Plasmids: Expression plasmids for wild-type SpCas9 and the high-fidelity variant (e.g., SpCas9-HF1).
- sgRNA Oligonucleotides: Designed for the endogenous locus of interest (e.g., EMX1, FANCF).
- Cells: HEK293T or other relevant human cell lines.
- GUIDE-seq dsODN: A 34-bp double-stranded oligodeoxynucleotide tag with phosphorothioate modifications (e.g., 5'-GTCTCTGATCGGCTACTAGCGTACGCATAGCCTGCAG-3', where * = phosphorothioate bond).
- Transfection Reagent: Lipofectamine 3000 or similar.
- Lysis Buffer: For genomic DNA extraction.
- PCR and NGS Reagents: Primers for on-target and potential off-target loci, next-generation sequencing library preparation kit.
Procedure
sgRNA Cloning: Clone the annealed sgRNA oligonucleotides into the respective Cas9 expression plasmid backbone using standard molecular biology techniques (e.g., BbsI restriction site digestion and ligation).
Cell Transfection:
- Culture HEK293T cells in appropriate medium (e.g., DMEM + 10% FBS) until 70-80% confluent.
- For each sample (WT Cas9 + sgRNA, High-Fidelity Cas9 + sgRNA), prepare a transfection mixture containing:
- 1 µg of Cas9-sgRNA plasmid
- 100 pmol of GUIDE-seq dsODN tag
- Transfect the cells according to the manufacturer's protocol for the transfection reagent.
- Include a negative control (cells transfected with a non-targeting sgRNA).
Genomic DNA Harvesting:
- 72 hours post-transfection, wash the cells with PBS and harvest them by trypsinization.
- Extract high-molecular-weight genomic DNA using a commercial kit. Quantify DNA concentration and assess purity.
GUIDE-seq Library Preparation and Sequencing:
- dsODN Tag Integration Enrichment: Perform a first-round PCR with primers specific to the dsODN tag to enrich for genomic regions that have incorporated the tag.
- Indexing PCR: Use the first PCR product as a template for a second, limited-cycle PCR to add Illumina sequencing adapters and sample-specific barcodes.
- Purify the final library and validate its quality (e.g., using a Bioanalyzer). Sequence on an Illumina platform (e.g., MiSeq) with 150 bp paired-end reads.
Data Analysis:
- Sequence Alignment: Process the raw sequencing reads to remove adapter sequences and low-quality bases. Align the cleaned reads to the reference human genome (e.g., hg38) using an aligner like BWA-MEM.
- DSB Site Identification: Use the GUIDE-seq analysis software (e.g., the "GUIDE-seq" R package) to identify significant peaks of dsODN tag integration, which correspond to Cas9-induced double-strand breaks.
- Off-Target Comparison: Compare the list of identified off-target sites between wild-type SpCas9 and the high-fidelity variant. A true high-fidelity variant should show a significant reduction or complete absence of off-target sites while maintaining a strong on-target signal.
The Scientist's Toolkit: Essential Reagents for High-Fidelity Editing
Table 3: Key Research Reagent Solutions for High-Fidelity CRISPR Experiments
| Reagent / Solution | Function and Importance | Example Source / Specification |
|---|---|---|
| High-Fidelity Cas9 Plasmids | Mammalian expression vectors encoding SpCas9-HF1, eSpCas9(1.1), or HypaCas9. Essential for delivering the engineered nuclease. | Available from Addgene (e.g., Plasmid #72247 for SpCas9-HF1). |
| GUIDE-seq dsODN Tag | A synthetic double-stranded oligodeoxynucleotide that integrates into Cas9-induced DSBs, enabling genome-wide off-target profiling. | 34-bp, HPLC-purified, with phosphorothioate modifications on the first five 5' and 5' 3' nucleotides for stability [96]. |
| T7 Endonuclease I (T7EI) | Detects indel mutations at the on-target site by cleaving heteroduplex DNA formed from heterogenous PCR products. A quick, cost-effective validation tool. | Commercial kits from suppliers like New England Biolabs or Integrated DNA Technologies. |
| Lipofectamine 3000 | A high-efficiency lipid nanoparticle-based transfection reagent for delivering plasmids and the GUIDE-seq tag into cultured human cells. | Thermo Fisher Scientific. |
| Next-Generation Sequencing (NGS) Library Prep Kit | For preparing sequencing libraries from GUIDE-seq PCR products or targeted amplicons to quantitatively assess editing efficiency and specificity. | Kits such as Illumina's Nextera XT. |
Clinical Relevance and Future Perspectives
The development of high-fidelity Cas9 variants is not merely an academic exercise; it is a critical step toward enabling safe and effective therapeutic applications of CRISPR genome editing. Off-target effects represent a major safety concern for clinical trials, and the use of more precise nucleases can mitigate this risk [9] [44].
The translation of CRISPR technology into the clinic is advancing rapidly. As of early 2025, the first CRISPR-based medicine, Casgevy, has been approved for sickle cell disease and transfusion-dependent beta thalassemia, and over 150 active clinical trials are underway for a wide range of conditions, including blood cancers, hereditary amyloidosis, and viral diseases [9] [80]. The integration of high-fidelity variants like SpCas9-HF1 into therapeutic pipelines is a logical progression. For instance, recent work has shown that SpCas9-HF1 can be successfully incorporated into advanced editing strategies, such as cell cycle-dependent genome editing, to further enhance Homology-Directed Repair (HDR) efficiency while minimizing off-target effects [99].
Looking forward, the combination of high-fidelity nucleases with advanced delivery systems, such as lipid nanoparticles (LNPs), promises to improve the safety and efficacy of in vivo genome editing [9] [44]. As the structural and mechanistic understanding of these engineered variants deepens, it will continue to inform the rational design of next-generation editors with even greater precision and versatility for both basic research and therapeutic development.
The CRISPR-Cas9 system has revolutionized genome editing by enabling precise, programmable modifications of DNA. However, its targeting capacity is constrained by a fundamental requirement: the presence of a short protospacer adjacent motif (PAM) sequence immediately adjacent to the target site [1]. For the widely used Streptococcus pyogenes Cas9 (SpCas9), this PAM is the 5'-NGG-3' sequence, where "N" can be any nucleotide [100]. This limitation restricts targeting to genomic regions flanked by NGG sites, which constitute only a fraction of the genome and may exclude therapeutically relevant loci [101]. To overcome this bottleneck, significant protein engineering efforts have yielded novel Cas9 variants with dramatically expanded PAM compatibility, including xCas9, SpCas9-NG, and SpRY, thereby increasing the accessible genomic landscape for therapeutic editing [102] [100].
Engineered Cas9 Variants with Expanded PAM Compatibility
xCas9: Flexibility-Driven PAM Recognition
xCas9, an engineered variant developed through directed evolution, incorporates seven amino acid substitutions (A262T, R324L, S409I, E480K, E543D, M694I, E1219V) that collectively broaden its PAM recognition capabilities [102]. Unlike SpCas9, which enforces strict guanine selection through a rigid arginine dyad (R1333 and R1335), xCas9 introduces structural flexibility that enables recognition of altered PAM sequences [102].
Key Mechanistic Insight: The primary mechanism for xCas9's expanded PAM recognition involves increased conformational flexibility at residue R1335. While SpCas9 maintains rigid arginine interactions that strictly select for guanine, xCas9's flexible R1335 enables selective recognition of diverse PAM sequences through an entropic preference that also enhances binding to the canonical TGG PAM [102]. The E1219V mutation within the PAM-interacting domain does not directly contact DNA but allosterically facilitates this flexibility, enabling recognition of both guanine and adenine-containing PAMs [102].
xCas9 demonstrates robust activity with PAM sequences including NGN (N= A, G, C, or T), with particularly high efficiency for GAG, GAA, GAT, and TGG [102]. The variant also exhibits reduced off-target effects compared to SpCas9, making it valuable for applications requiring high specificity [102].
SpCas9-NG: Relaxing the Third Position Constraint
SpCas9-NG represents another engineered variant designed to overcome PAM restrictions by relaxing the stringency at the third position of the PAM sequence [100]. Through targeted mutations (R1335V/L1111R/D1135V/G1218R/E1219F/A1322R/T1337R), SpCas9-NG recognizes NG PAMs, where the third nucleotide can be any base, significantly expanding the targeting range compared to wild-type SpCas9 [100].
While SpCas9-NG substantially increases targetable sites, its editing efficiency varies considerably across different NG PAMs, with reported higher activity for PAMs ending in G or T, and lower efficiency for those ending in C [100]. This variant maintains robust on-target activity while reducing off-target effects in certain contexts, though its performance is highly dependent on the specific PAM sequence and target locus [100].
SpRY: Nearly PAMless Editing
SpRY represents the most flexible engineered Cas9 variant, approaching a "PAMless" editing capability [101] [100]. Through comprehensive engineering of the PAM-interacting domain (L1111R, D1135L, S1136W, G1218K, E1219Q, A1322R, R1333P, R1335Q, T1337R), SpRY effectively recognizes NRN and NYN PAMs, where R is A/G and Y is C/T [101] [100]. This broad specificity enables targeting of virtually any genomic sequence, with a preference for NRN > NYN PAMs [101].
In human cells, SpRY demonstrates efficient editing across diverse PAM sequences, with canonical PAMs identified as NRN, NTA, and NCK, while NCA and NTK serve as non-priority PAMs [100]. The 5'-NYC-3' PAM is not recommended due to minimal activity [100]. SpRY has been successfully deployed for targeting previously inaccessible disease-relevant mutations, including specific pathogenic SNPs that require non-canonical PAM recognition [101].
SpRYc: A Chimeric Solution
Building on SpRY's flexibility, researchers developed SpRYc, a chimeric enzyme that combines the N-terminus of Sc++ (including its positive-charged loop structure) with the PAM-interacting domain of SpRY [101]. This hybrid architecture integrates the broad PAM compatibility of SpRY with the efficient NNG editing capability of Sc++ [101].
SpRYc exhibits highly flexible PAM preference, enabling efficient base editing at genomic sites with minimal PAM dependence [101]. When fused to the ABE8e adenine base editor, SpRYc outperforms both SpCas9-ABE8e and SpRY-ABE8e on certain PAMs, particularly 5'-NTN-3' and 5'-NNT-3' sequences [101]. Notably, SpRYc demonstrates nearly four-fold lower off-target activity than SpRY at the VEGFA site and two-fold lower off-targeting at EMX1, suggesting improved specificity alongside its broad targeting range [101].
Table 1: Comparison of PAM-Flexible Cas9 Variants
| Variant | Key Mutations | PAM Preference | Editing Efficiency | Primary Applications |
|---|---|---|---|---|
| xCas9 | A262T, R324L, S409I, E480K, E543D, M694I, E1219V | NGN (particularly GAN) | Variable across PAMs; high for GAG, GAA, GAT | Applications requiring reduced off-target effects with moderate PAM flexibility |
| SpCas9-NG | R1335V/L1111R/D1135V/G1218R/E1219F/A1322R/T1337R | NG | Higher for NGG, NGT; lower for NGC, NGA | Targeting sites with NG PAMs where canonical NGG is unavailable |
| SpRY | L1111R, D1135L, S1136W, G1218K, E1219Q, A1322R, R1333P, R1335Q, T1337R | NRN > NYN (near PAMless) | Broad but with preference for NRN | Maximizing targetable space; accessing previously inaccessible disease loci |
| SpRYc | Chimeric fusion of SpRY PID with Sc++ N-terminus | NRN ~ NYN (highly flexible) | High across diverse PAMs; outperforms SpRY on NTN/NNT | Therapeutic applications requiring precise positioning with minimal PAM constraints |
Experimental Methodologies for PAM Characterization
PAM-SCANR: A Bacterial Selection System
The PAM-SCANR (PAM Definition by Observable Sequence Excision) system represents a bacterial-based approach for functional PAM determination [100]. This method employs a positive selection system where cell survival or reporter expression depends on functional PAM recognition and subsequent DNA cleavage.
Protocol:
- Clone a randomized PAM library (typically 5'-NNN-3') into a plasmid vector upstream of a fixed protospacer sequence adjacent to a selectable or screenable marker [100].
- Co-transform the PAM library plasmid with plasmids expressing the Cas9 variant of interest and a corresponding sgRNA targeting the fixed protospacer [100].
- Apply selection pressure (e.g., antibiotic resistance) or screen for reporter expression (e.g., GFP) to isolate cells with functional PAM sequences [100].
- Isolate plasmids from surviving cells and sequence the PAM region to determine which sequences support functional Cas9 activity [100].
This method provides a rapid, high-throughput approach for initial PAM characterization, though it may not fully recapitulate the chromatin environment and cellular factors present in eukaryotic systems [100].
HT-PAMDA: High-Throughput PAM Determination Assay
HT-PAMDA represents a more quantitative, in vitro approach for comprehensive PAM characterization [101]. This method directly measures the cleavage kinetics of Cas enzymes on a library of DNA substrates containing randomized PAM sequences.
Protocol:
- Generate a DNA library containing a randomized PAM region (e.g., 5'-NNN-3') flanking a fixed target sequence [101].
- Incubate the library with the Cas9 variant and corresponding sgRNA under defined reaction conditions [101].
- Isolate and sequence the cleaved products over multiple timepoints to determine cleavage rates for each PAM sequence [101].
- Calculate relative cleavage efficiencies by comparing the enrichment or depletion of specific PAM sequences in the cleaved fraction versus the input library [101].
HT-PAMDA provides quantitative kinetic data rather than binary functional assessment, enabling more nuanced understanding of PAM preference and efficiency [101]. This approach was used to characterize SpRYc, revealing broader editing capabilities than SpCas9 and Sc++, though with somewhat slower cleavage rates than SpRY [101].
Cell-Based PAM Validation: GFP-Reporter and PAM-DOSE
For validation in human cells, the GFP-reporter system and PAM-DOSE (PAM Definition by Observable Sequence Excision) enable functional assessment of PAM compatibility in a more physiologically relevant context [100].
GFP-Reporter Protocol:
- Engineer a reporter construct containing a GFP gene interrupted by a Cas9 target site with a specific PAM sequence [100].
- Co-transfect cells with the reporter construct along with plasmids expressing the Cas9 variant and sgRNA [100].
- Quantify GFP-positive cells via flow cytometry; successful editing results in GFP expression restoration [100].
- Compare editing efficiencies across different PAM sequences by normalizing GFP-positive percentages to control conditions [100].
PAM-DOSE Protocol:
- Create a library of target sites containing randomized PAM sequences in a reporter construct [100].
- Introduce the library into human cells (e.g., HEK293) along with Cas9 and sgRNA expression vectors [100].
- Isolate and sequence the target regions from successfully edited cells to determine functional PAM preferences [100].
These cell-based approaches confirmed that SpRY recognizes 5'-NRN-3', 5'-NTA-3', and 5'-NCK-3' as canonical PAMs in human cells, with 5'-NCA-3' and 5'-NTK-3' as non-priority PAMs, while 5'-NYC-3' is not recommended [100].
Genome-Wide Off-Target Assessment
Comprehensive characterization of PAM-flexible variants requires rigorous off-target profiling. GUIDE-Seq provides a genome-wide, unbiased method for identifying off-target sites [101].
GUIDE-Seq Protocol:
- Transfect cells with Cas9/sgRNA along with blunt-ended, double-stranded oligodeoxynucleotides (dsODNs) that serve as tags for double-strand break sites [101].
- Harvest genomic DNA and shear by sonication [101].
- Prepare sequencing libraries using adaptors compatible with high-throughput sequencing platforms [101].
- Enrich for dsODN-integrated fragments and sequence [101].
- Map reads to the reference genome to identify off-target sites, comparing to negative controls [101].
Using GUIDE-Seq, researchers demonstrated that SpRYc exhibits nearly four-fold lower off-target activity than SpRY at the VEGFA site and two-fold lower off-targeting at EMX1, suggesting its chimeric architecture may provide improved specificity [101].
Research Reagent Solutions
Table 2: Essential Research Reagents for PAM-Flexible Cas9 Studies
| Reagent/Tool | Function | Example Application | Key Features |
|---|---|---|---|
| PAM-SCANR System | Functional PAM determination in bacteria | Initial screening of PAM preferences | Rapid, high-throughput bacterial selection system [100] |
| HT-PAMDA Library | Quantitative PAM characterization in vitro | Kinetic analysis of cleavage efficiency | Provides quantitative data on cleavage rates across PAMs [101] |
| GFP-Reporter Constructs | PAM validation in human cells | Confirming functional PAMs in physiological context | Enables flow cytometry-based quantification [100] |
| GUIDE-Seq | Genome-wide off-target profiling | Assessing specificity of PAM-flexible variants | Unbiased identification of off-target sites [101] |
| qEva-CRISPR | Quantitative editing efficiency measurement | Multiplex analysis of target and off-target sites | Detects all mutation types; suitable for difficult genomic regions [103] |
| CRISPRainbow System | Multiplexed genomic locus visualization | Tracking chromosomal dynamics and editing outcomes | Enables 6-color imaging of genomic loci [104] |
The engineering of PAM-flexible Cas9 variants represents a significant advancement in CRISPR-based technologies, dramatically expanding the targetable genomic landscape for research and therapeutic applications. xCas9, SpCas9-NG, SpRY, and the chimeric SpRYc each offer distinct advantages depending on the specific editing requirements, balancing PAM flexibility with editing efficiency and specificity [101] [100] [102]. These tools enable targeting of previously inaccessible disease-relevant loci, including specific mutations in genes such as MECP2 for Rett syndrome that require non-canonical PAM recognition [101]. As the field progresses, continued refinement of these enzymes—optimizing both their targeting scope and specificity—will further enhance their utility for both basic research and clinical applications, ultimately expanding the therapeutic potential of genome editing for addressing genetic diseases.
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9 system has emerged as a revolutionary technology for genome editing, with transformative potential for both basic research and therapeutic applications. Its core principle involves a guide RNA (gRNA) directing the Cas9 nuclease to a specific genomic locus, where it introduces a double-strand break (DSB) that is subsequently repaired by endogenous cellular machinery [50] [16]. However, the transition from a powerful in vitro tool to a reliable in vivo therapeutic hinges on overcoming three interconnected delivery challenges: achieving tissue-specific targeting, evading the host immune system, and circumventing stringent cargo size limitations. This guide examines the principles and advanced strategies addressing these hurdles, providing a framework for optimizing CRISPR-Cas9 delivery within the broader context of genome editing research.
Cargo Size Constraints and Engineering Solutions
The efficient packaging of CRISPR components is a primary logistical challenge. The most commonly used Cas9 nuclease from Streptococcus pyogenes (SpCas9) has a coding sequence of approximately 4.2 kb, which, when combined with promoter elements and gRNA sequences, easily exceeds the packaging capacity of popular viral vectors like adeno-associated virus (AAV), limited to ~4.7 kb [50] [49].
Strategies to Overcome Size Limitations
Table 1: Strategies for Delivering Large CRISPR Cargos
| Strategy | Mechanism | Key Examples | Advantages | Limitations |
|---|---|---|---|---|
| Compact Cas Orthologs | Use of naturally smaller Cas proteins. | SaCas9 (∼3.2 kb), CjCas9 (∼3.0 kb), Cas12f (∼1.5-2 kb) [49] | Fits into a single AAV vector with space for promoters/gRNA. | May have different PAM requirements or lower efficiency than SpCas9. |
| Dual Vector Systems | Splitting CRISPR components across two AAV vectors. | One vector for Cas9, another for gRNA(s) and donor template [105] [49]. | Delivers full-length SpCas9 and complex expression cassettes. | Lower effective co-infection rate; potential for incomplete editing. |
| Ribonucleoprotein (RNP) Delivery | Direct delivery of pre-assembled Cas9 protein and gRNA complex. | Cas9 protein complexed with sgRNA [105] [106]. | Immediate activity, reduced off-target effects, transient presence. | Requires efficient in vivo delivery methods (e.g., nanoparticles). |
| Virus-Like Particles (VLPs) | Engineered viral capsids lacking viral genetic material. | VLP packaged with Cas9 RNP [105]. | Transient delivery, high safety profile, avoids DNA integration. | Manufacturing challenges and cargo size limitations persist. |
Protein Engineering for Expanded PAM Recognition
Beyond size, the Protospacer Adjacent Motif (PAM) sequence required for Cas9 target recognition restricts the editable genomic landscape. Protein engineering has created "PAM-flexible" Cas9 variants like SpRY, which recognizes NRN (R = A/G) and NYN (Y = C/T) PAMs, vastly increasing targetable sites [16]. Engineering efforts, informed by molecular dynamics simulations, show that effective PAM recognition involves not only direct DNA-contact residues but also a distal network that stabilizes the PAM-binding domain and preserves long-range allosteric communication within the Cas9 protein [107].
Tissue and Organ-Specific Targeting Strategies
Effective in vivo therapy requires the CRISPR machinery to reach the nucleus of the correct cell type. This is confounded by numerous physiological barriers, including blood clearance, vascular endothelium, and the reticuloendothelial system [66].
Viral Vector Engineering
Recombinant AAV (rAAV) vectors are prominent for in vivo delivery due to their low immunogenicity and high tissue specificity. Different AAV serotypes (e.g., AAV8 for liver, AAV9 for heart and CNS, AAV5 for retina) exhibit natural tropism for specific tissues [49]. This tropism can be further enhanced by engineering the viral capsid to display peptides that bind to receptors uniquely expressed on target cell surfaces, a process known as "directed evolution" [66] [49].
Non-Viral Vector Engineering
Non-viral vectors, particularly Lipid Nanoparticles (LNPs), have gained prominence for their organ-targeting potential.
- Selective Organ Targeting (SORT): A key advancement involves engineering LNPs with additional "SORT" molecules. The lipid composition determines in vivo destination; for instance, ionizable lipids with permanent positive charge tend to accumulate in the lung, while those with a pKa between 6.0 and 6.5 preferentially target the liver [105] [66].
- Extracellular Vesicles (EVs): EVs are natural lipid nanoparticles derived from cells, inherently possessing homing capabilities to specific tissues. They offer high biocompatibility and can be engineered to display targeting motifs on their surface [50] [105].
The following diagram illustrates the two primary engineering approaches for achieving tissue-specific delivery.
Immune Evasion and Safety Optimization
The host immune system can mount responses against both the delivery vector and the bacterial-derived Cas9 protein, potentially reducing efficacy and causing toxicity.
Mitigating Immune Recognition
Table 2: Immune Evasion and Safety Strategies
| Challenge | Strategy | Mechanism | Reference |
|---|---|---|---|
| Preexisting Immunity to AAV | Use of rare human or synthetic AAV capsids. | Avoids neutralization by preexisting antibodies common in the human population. | [49] |
| Adaptive Immunity to Cas9 | Use of transient delivery formats (RNP, mRNA). | Limits sustained Cas9 exposure, reducing T-cell activation and immune memory. | [105] |
| Cytotoxicity from Persistent Expression | Delivery as Ribonucleoprotein (RNP). | The protein-RNA complex is active for only a short period, minimizing off-target effects. | [105] [106] |
| Off-Target Editing | Use of high-fidelity Cas9 variants; RNP delivery. | eSpCas9(1.1), SpCas9-HF1, HypaCas9 reduce off-target activity by altering DNA binding. RNP's transient activity also helps. | [16] [108] |
Advanced Detection of Off-Target Effects
Ensuring safety requires robust methods to identify unintended genomic edits. Moving beyond in silico prediction, several unbiased, genome-wide screening methods have been developed:
- GUIDE-seq: Uses a short, double-stranded oligonucleotide tag that integrates into DSBs, allowing for their precise sequencing-based identification [108].
- Digenome-seq: Involves cleaving purified genomic DNA with Cas9 RNP in vitro, followed by whole-genome sequencing to map all cleavage sites [108].
- BLESS: Directly ligates sequencing adapters to DSBs in fixed cells, capturing breaks in situ [108].
The Scientist's Toolkit: Essential Reagents and Methods
This section details key experimental reagents and protocols central to advanced CRISPR delivery research.
Research Reagent Solutions
Table 3: Key Reagents for CRISPR Delivery Optimization
| Reagent / Material | Function in Delivery Optimization | Example Application |
|---|---|---|
| High-Fidelity Cas9 (e.g., eSpCas9) | Engineered protein with reduced off-target effects. | Critical for therapeutic applications where specificity is paramount. [16] [108] |
| Compact Cas Orthologs (e.g., SaCas9) | Enables packaging into single AAV vector. | In vivo gene therapy where AAV is the preferred vector. [49] |
| Chemically Modified gRNA | Increases gRNA stability and reduces immunogenicity. | Used with RNP or LNP delivery to enhance efficiency and safety. [106] |
| Ionizable Lipid Nanoparticles | Forms the core of non-viral delivery vehicles for mRNA/RNP. | Systemic in vivo delivery to organs like the liver. [105] [66] |
| ssODN Donor Template | Serves as a repair template for precise HDR-mediated editing. | Introducing specific point mutations or small insertions. [106] |
Experimental Protocol: HDR with ssODN Donor Templates
Homology-Directed Repair (HDR) allows for precise genome editing but is less efficient than error-prone Non-Homologous End Joining (NHEJ). The following workflow, optimized for RNP delivery, maximizes HDR efficiency [106]:
Key design parameters for the ssODN template [106]:
- Homology Arm Length: 30-40 nucleotides on each side is often optimal.
- Strand Preference: The choice between using the targeting (T) or non-targeting (NT) strand as the donor template is locus-dependent and should be determined empirically.
- Blocking Mutations: Incorporate silent mutations into the PAM sequence in the donor template to prevent Cas9 from re-cleaving the successfully edited locus.
The journey of CRISPR-Cas9 from a laboratory tool to a therapeutic modality is intrinsically linked to solving the tripartite challenge of delivery: cargo, targeting, and stealth. Innovations in protein engineering have yielded smaller, more precise Cas variants and expanded the targetable genome. Simultaneously, vector engineering, through both viral capsid and synthetic nanoparticle design, is paving the way for true tissue-specific tropism. Underpinning these advances is a growing emphasis on safety, driven by transient delivery methods like RNP and sophisticated in vivo immune evasion strategies. The integration of artificial intelligence and machine learning promises to further accelerate the design of novel editors and delivery systems with enhanced precision and functionality [50] [66]. As these technologies mature, the systematic optimization of delivery vectors will continue to be a cornerstone of basic CRISPR research and its translation into transformative gene therapies.
The Clustered Regularly Interspaced Short Palindromic Repeats and associated Cas9 protein (CRISPR-Cas9) system has revolutionized genome engineering, unlocking unprecedented therapeutic potential for genetic diseases, cancer, and beyond [84]. Derived from a bacterial adaptive immune system, this technology functions as a programmable gene-editing tool capable of making precise changes to genomic DNA [109]. The system operates through a simple yet powerful mechanism: a Cas nuclease, directed by a guide RNA (gRNA), recognizes a target DNA sequence via Watson-Crick base pairing and induces a double-strand break (DSB) [27]. This break activates the cellular DNA damage response, leading to both intended and unintended genetic modifications through endogenous repair pathways [27].
As CRISPR-based therapies progress toward clinical application—exemplified by the recent regulatory approval of exa-cel (Casgevy) for sickle cell disease and transfusion-dependent beta thalassemia—comprehensive safety assessments have become paramount [27] [9]. Beyond well-documented concerns about off-target mutagenesis, emerging evidence reveals more pressing challenges: large structural variations and immune recognition of bacterial-derived Cas proteins [27] [110]. These undervalued genomic alterations and immunological responses raise substantial safety concerns for clinical translation, requiring rigorous evaluation in pre-clinical studies and continued monitoring in clinical trials [111]. This technical guide examines these critical safety considerations within the broader context of basic principles of CRISPR-Cas9 genome editing research, providing researchers and drug development professionals with a comprehensive framework for risk assessment and mitigation.
Immune Responses to CRISPR-Cas9 Components
Mechanisms of Immunogenicity
The immunogenicity of CRISPR-Cas9 components presents a significant challenge for in vivo therapies. Immune recognition can trigger both innate and adaptive responses, potentially impacting both the safety and efficacy of CRISPR-based treatments [110]. The complex interactions between Cas9, delivery vectors, and host immune reactivity play a crucial role in determining clinical outcomes.
Pre-existing immunity to Cas proteins is common in human populations, with approximately 80% of people showing immunological exposure to these bacterial-derived proteins through natural environmental exposure [112]. This immunity arises because the Cas9 nuclease commonly used in CRISPR applications is derived from Streptococcus pyogenes and Staphylococcus aureus, bacteria that frequently colonize or infect humans [109]. When administered in vivo, these bacterial proteins can be recognized by the host immune system, potentially leading to reduced therapy efficacy due to rapid clearance of edited cells or triggering inflammatory responses that pose safety risks [110].
Epitope Identification and Engineering Solutions
Recent research has made significant strides in characterizing the specific components of Cas proteins that trigger immune responses. Using specialized mass spectrometry techniques, researchers have identified immunogenic sequences within both Cas9 and Cas12 proteins [112]. For Cas9 from Streptococcus pyogenes and Cas12 from Staphylococcus aureus, three short sequences approximately eight amino acids long each were found to evoke immune responses [112].
This structural knowledge has enabled rational protein engineering to create minimized immunogenicity variants. Through computational modeling and structure-based design approaches, researchers have developed engineered Cas enzymes with modified surface epitopes that reduce immune recognition while maintaining editing functionality [112]. Studies in humanized mouse models have demonstrated that these engineered nucleases exhibit significantly reduced immune responses while preserving DNA cleavage efficiency comparable to their wild-type counterparts [112].
Table 1: Engineered CRISPR Nucleases with Reduced Immunogenicity
| Nuclease Name | Parent Protein | Engineering Approach | Immune Response Reduction | Editing Efficiency |
|---|---|---|---|---|
| engineered Cas9 | S. pyogenes Cas9 | Epitope masking via computational design | Significant reduction in humanized mice | Equivalent to wild-type |
| engineered Cas12 | S. aureus Cas12 | Rational engineering of immune-triggering sequences | Significantly reduced immune recognition | Maintained target cleavage |
| hpABE5.20 | ABE8e base editor | Precision engineering for reduced off-target effects | N/A (reduced off-target editing) | Matched ABE8e with improved precision |
Experimental Protocols for Immunogenicity Assessment
Protocol 1: In Vitro Immunogenicity Screening
- Isolate human peripheral blood mononuclear cells (PBMCs) from healthy donors using density gradient centrifugation.
- Culture PBMCs in RPMI-1640 medium supplemented with 10% fetal bovine serum.
- Prepare Cas9 protein in increasing concentrations (0.1-10 μg/mL).
- Stimulate PBMCs with Cas9 proteins for 24-48 hours.
- Measure T-cell activation via flow cytometry analysis of CD4+ and CD8+ T cells expressing CD69 and CD25.
- Quantify cytokine secretion using ELISA for IFN-γ, IL-2, and TNF-α in culture supernatants.
- Compare response to positive (anti-CD3/CD28 antibodies) and negative (media alone) controls.
Protocol 2: In Vivo Immunogenicity Assessment in Humanized Mouse Models
- Engineer immunodeficient mice (e.g., NSG strains) to express key components of the human immune system.
- Administer CRISPR components via the intended therapeutic delivery route (LNP, AAV, etc.).
- Collect blood samples at predetermined intervals (days 1, 3, 7, 14, 28 post-administration).
- Analyze serum for anti-Cas antibodies using enzyme-linked immunosorbent assay (ELISA).
- Isolate splenocytes at endpoint for T-cell recall assays by re-stimulating with Cas9 peptides.
- Evaluate cellular immune responses via interferon-γ ELISpot and intracellular cytokine staining.
- Assess inflammatory markers and tissue damage through histopathology of major organs.
Unintended Genomic Consequences
Spectrum of On-Target Structural Variations
While early CRISPR safety assessments primarily focused on off-target effects at sites with sequence similarity to the intended target, recent evidence reveals a more complex landscape of unintended on-target outcomes extending beyond simple insertions or deletions (indels) [27]. These include:
Kilobase- to megabase-scale deletions at the on-target site that can remove extensive genomic regions, potentially eliminating critical genes and regulatory elements [27]. Studies targeting the BCL11A enhancer in hematopoietic stem cells (HSCs) for sickle cell disease therapy have demonstrated these large deletions occurring frequently, warranting closer scrutiny despite successful clinical outcomes [27].
Chromosomal rearrangements including translocations between homologous chromosomes that result in an acentric and a dicentric chromosome, large deletions following two cleavage events on the same chromosome, and translocations between different heterologous chromosomes upon simultaneous cleavage of the target site and an off-target site [27]. These rearrangements are particularly concerning as they can activate oncogenes or inactivate tumor suppressors.
Chromothripsis, a catastrophic event where chromosomes undergo massive shattering and rearrangement in a single event, has been observed in some CRISPR-edited cells [27]. This phenomenon is particularly associated with p53-deficient cells and has significant implications for cancer risk assessment in therapeutic editing.
Notably, although these genomic alterations have been more extensively studied in the context of the CRISPR/Cas system, similar effects have also been observed with other DSB-inducing platforms, such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) [27].
Impact of DNA Repair Pathway Manipulations
The cellular response to CRISPR-induced double-strand breaks is mediated primarily by two competing repair pathways: non-homologous end joining (NHEJ) and homology-directed repair (HDR) [27] [84]. The choice between these pathways has profound implications for both editing outcomes and genomic integrity.
Researchers have developed strategies to enhance HDR efficiency for precise editing, often through chemical inhibition of key NHEJ pathway components. However, recent findings reveal that these approaches can inadvertently introduce new risks. The use of DNA-PKcs inhibitors such as AZD7648—frequently adopted for promoting HDR by suppressing NHEJ—can lead to exacerbated genomic aberrations [27]. These compounds significantly increase frequencies of kilobase- and megabase-scale deletions as well as chromosomal arm losses across multiple human cell types and loci [27].
Furthermore, off-target profiles are markedly aggravated with DNA-PKcs inhibition, with surveys of off-target-mediated chromosomal translocations revealing not only a qualitative rise in the number of translocation sites but also an alarming thousand-fold increase in the frequency of such structural variations [27]. These findings align with other studies investigating the impact of alternative DNA-PKcs inhibitors, confirming that disturbing the NHEJ repair pathway alters the genomic landscape in unpredictable ways [27].
Table 2: Unintended Genomic Consequences of CRISPR-Cas9 Editing
| Genomic Alteration | Size Range | Detection Methods | Reported Frequency | Risk Factors |
|---|---|---|---|---|
| Small indels | 1-100 bp | Amplicon sequencing, NGS | Variable (5-90%) | NHEJ preference, gRNA design |
| Kilobase-scale deletions | 1-100 kb | CAST-Seq, LAM-HTGTS, long-read sequencing | Up to 15% in HSCs | DNA-PKcs inhibition, high nuclease concentration |
| Megabase-scale deletions | >100 kb | CAST-Seq, LAM-HTGTS, karyotyping | ~10% in some loci | DNA-PKcs inhibition, p53 deficiency |
| Chromosomal translocations | N/A | CAST-Seq, LAM-HTGTS, cytogenetics | Up to 1000× increase with NHEJ inhibition | Simultaneous DSBs, DNA-PKcs inhibition |
| Chromothripsis | Chromosome-scale | Karyotyping, whole-genome sequencing | Rare | p53 deficiency, cell cycle dysregulation |
Methodological Limitations in Outcome Detection
Traditional analytical approaches for assessing editing outcomes may significantly underestimate the prevalence of large-scale structural variations. Short-read amplicon sequencing techniques, commonly used to quantify editing efficiency, fail to detect extensive deletions or genomic rearrangements that delete primer-binding sites, rendering these events 'invisible' to analysis [27]. This methodological limitation has led to systematic overestimation of HDR rates and concurrent underestimation of indels and structural variations in many studies [27].
Advanced detection methods have been developed to comprehensively assess these genomic alterations, including:
CAST-Seq (CRISPR Antiviral Defense Screening by Sequencing) - specifically designed to detect structural variations and translocations resulting from CRISPR editing [27].
LAM-HTGTS - a high-throughput genome-wide translocation sequencing approach that maps chromosomal rearrangements genome-wide [27].
Long-read sequencing (Oxford Nanopore, PacBio) - enables detection of large structural variations that span beyond the capacity of short-read technologies [27].
These methodologies have revealed that the genomic consequences of CRISPR editing are substantially more complex than initially appreciated, emphasizing the need for comprehensive genotoxicity assessment in therapeutic development.
Experimental Protocols for Safety Assessment
Comprehensive Structural Variation Analysis
Protocol 1: CAST-Seq for Translocation Detection
- Design target-specific biotinylated primers flanking the CRISPR target site(s).
- Extract high-molecular-weight genomic DNA from edited cells using gentle extraction methods to prevent DNA shearing.
- Perform linker-adapter PCR to amplify regions of interest and potential translocation partners.
- Capture biotinylated products using streptavidin beads and prepare sequencing libraries.
- Sequence on appropriate platform (Illumina recommended for high coverage).
- Bioinformatic analysis: Map reads to reference genome, identify chimeric reads spanning breakpoints, and filter valid translocation events from artifacts.
- Validate high-frequency translocations by orthogonal methods such as PCR and Sanger sequencing.
Protocol 2: Long-read Sequencing for Large Structural Variations
- Extract high-quality genomic DNA using protocols optimized for long fragment preservation (e.g., magnetic bead-based systems).
- Quantity DNA yield and purity using fluorometric methods and assess integrity via pulsed-field or standard gel electrophoresis.
- Prepare sequencing libraries according to platform-specific protocols (Oxford Nanopore or PacBio).
- Sequence with sufficient coverage (typically 20-30× for variant detection).
- Perform structural variant calling using specialized algorithms (e.g., Sniffles, PBSV).
- Annotate variants with gene features and regulatory elements to assess potential functional impact.
- Prioritize variants based on size, gene content, and potential pathogenicity for further validation.
Experimental Design for Risk Mitigation
To systematically evaluate and mitigate the risks of unintended genomic consequences, the following experimental approaches are recommended:
Dose-response studies to determine the minimum editor concentration/duration required for therapeutic efficacy, as higher doses correlate with increased structural variations [27].
Time-course analyses to assess the stability of genomic alterations over multiple cell divisions, as some abnormalities may be eliminated through selective pressures.
Multiple gRNA testing to identify guides with optimal efficiency-to-safety ratios, as gRNA sequence and chromatin context influence both on-target and off-target effects.
Repair pathway modulation with careful consideration of potential consequences, as strategies to enhance HDR (e.g., NHEJ inhibition) may exacerbate structural variations while improving precise editing rates [27].
Research Reagent Solutions
Table 3: Essential Research Reagents for CRISPR Safety Assessment
| Reagent/Category | Specific Examples | Function/Application | Safety Consideration Addressed |
|---|---|---|---|
| High-fidelity Cas variants | HiFi Cas9, Cas12a | Reduced off-target activity while maintaining on-target efficiency | Off-target effects |
| Immune-engineered nucleases | engineered Cas9, engineered Cas12 | Reduced immunogenicity for in vivo applications | Immune recognition |
| DNA repair inhibitors | AZD7648 (DNA-PKcs inhibitor), pifithrin-α (p53 inhibitor) | Modulate DNA repair pathway choice; enhance HDR | On-target structural variations |
| Advanced detection assays | CAST-Seq, LAM-HTGTS, long-read sequencing | Comprehensive identification of structural variations | Unintended genomic consequences |
| Delivery systems | LNPs, AAVs, electroporation systems | Efficient delivery with reduced immunogenicity | Immune responses, editing efficiency |
| Bioinformatics tools | CRISPResso, CHOPCHOP, Cas-OFFinder | gRNA design, efficiency prediction, outcome analysis | Multiple safety parameters |
| Cell viability assays | p53 pathway activation tests, apoptosis assays | Assessment of cellular stress responses | Genotoxicity |
| Control gRNAs | Non-targeting guides, targeting safe harbor loci | Experimental controls for specificity assessment | Benchmarking safety profiles |
The transformative potential of CRISPR-Cas9 genome editing in therapeutic applications is tempered by significant safety considerations involving immune responses and unintended genomic consequences. Pre-existing immunity to bacterial-derived Cas proteins poses challenges for in vivo therapies, potentially impacting both efficacy and safety. Meanwhile, advanced detection methods have revealed a previously underappreciated landscape of structural variations, including large deletions and chromosomal translocations, that extend beyond simple off-target effects.
Addressing these challenges requires integrated approaches combining protein engineering to reduce immunogenicity, advanced analytical methods to comprehensively assess genomic integrity, and careful modulation of DNA repair pathways. The recent development of engineered Cas enzymes with reduced immune recognition and improved specificity represents significant progress toward safer genome editing. Furthermore, recognizing that strategies to enhance precision editing, such as NHEJ inhibition, may inadvertently introduce new risks underscores the need for balanced therapeutic optimization.
As the field progresses toward more widespread clinical application, robust safety assessment frameworks must evolve in parallel. This includes standardized genotoxicity testing, comprehensive immune profiling, and long-term monitoring of edited cell populations. Through continued refinement of both editor design and safety evaluation methodologies, the field can work toward realizing the full therapeutic potential of CRISPR-based genome editing while effectively managing associated risks.
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated protein 9 (Cas9) system has revolutionized genome editing by providing researchers with an unprecedented ability to modify DNA sequences with high precision. This bacterial adaptive immune system has been engineered to function in eukaryotic cells, enabling targeted genetic alterations across a wide range of organisms and cell types [113]. At the core of this technology lies a simple yet powerful mechanism: the Cas9 nuclease is directed to a specific genomic location by a guide RNA (gRNA) that complementary base-pairs with the target DNA sequence adjacent to a short protospacer adjacent motif (PAM) [114].
The fundamental challenge in CRISPR-Cas9 genome editing lies in designing gRNAs that maximize on-target efficiency while minimizing off-target effects. An ideal gRNA must guide the Cas9 complex to the intended genomic locus with high specificity, avoiding unintended cleavage at similar sequences elsewhere in the genome [108] [114]. The design process has evolved from simple sequence alignment to sophisticated algorithmic approaches that incorporate machine learning models trained on large-scale experimental data. These advances have significantly improved the success rate of CRISPR experiments, making accurate gRNA design a critical first step in any genome editing workflow [115] [116].
Fundamental Principles of gRNA-Mediated Target Recognition
The CRISPR-Cas9 system recognizes target DNA through a dual recognition mechanism that requires both Watson-Crick base pairing between the gRNA and target DNA, and the presence of a specific PAM sequence immediately downstream of the target site [114]. For the most commonly used Streptococcus pyogenes Cas9 (SpCas9), the PAM sequence is 5'-NGG-3', where "N" can be any nucleotide [116]. This PAM requirement is a key determinant of target site selection and contributes to the specificity of the system by limiting potential target sites to those followed by this short motif.
Target recognition follows a two-step mechanism that is crucial for understanding both efficiency and specificity. Recent structural and biochemical studies reveal that Cas9 first engages DNA through non-specific interactions, followed by rapid PAM scanning. Once a compatible PAM is identified, Cas9 initiates DNA unwinding and permits gRNA-DNA hybridization [117]. This process begins at the PAM-distal end (seed region) and proceeds directionally toward the PAM. The stability of this R-loop formation - where the DNA strands are separated and the target strand base-pairs with the gRNA - directly determines cleavage efficiency [118].
Reduced PAM specificity has been shown to cause persistent non-selective DNA binding and recurrent failures to engage the target sequence through stable guide RNA hybridization, ultimately leading to reduced genome-editing efficiency in cells [117]. This reveals a fundamental trade-off between broad PAM recognition and genome-editing effectiveness, highlighting the importance of optimized target capture kinetics.
Table 1: Key Components of the CRISPR-Cas9 System and Their Functions
| Component | Structure/Composition | Function in Genome Editing |
|---|---|---|
| Cas9 Nuclease | Multi-domain protein with HNH and RuvC nuclease domains | Creates double-strand breaks in target DNA after specific recognition |
| Guide RNA (gRNA) | Synthetic fusion of crRNA and tracrRNA (single-guide RNA) | Directs Cas9 to specific genomic loci through complementary base pairing |
| Protospacer Adjacent Motif (PAM) | Short DNA sequence (e.g., NGG for SpCas9) | Essential for self/non-self discrimination; initiates Cas9 binding |
| Target DNA | 20-nucleotide sequence immediately upstream of PAM | Genomic locus to be edited; must be complementary to gRNA spacer |
Computational Approaches for Predicting On-Target Efficiency
On-target efficiency refers to the ability of a gRNA to direct Cas9 cleavage at its intended target site. Various computational approaches have been developed to predict this efficiency, evolving from simple rule-based methods to sophisticated machine learning models [114] [116]. These tools analyze sequence features that correlate with effective editing, allowing researchers to select optimal gRNAs before experimental validation.
The development of predictive models has been enabled by large-scale screens that measure the activity of thousands of gRNAs. Initial hypothesis-driven approaches identified key sequence features that influence efficiency, including GC content (optimal between 40-60%), specific nucleotide preferences at particular positions, and the avoidance of poly-N sequences (especially GGGG) [114] [119]. For example, guanines are strongly preferred at positions -1 and -2 relative to the PAM, while cytosines are favored at position 18 within the spacer [119].
More recently, learning-based approaches have surpassed rule-based methods in predictive accuracy. Conventional machine learning models like logistic regression and gradient boosting trees have been widely adopted, with deep learning methods now showing promising results [114]. These models automatically extract relevant features from gRNA sequences and their genomic context, often considering a 60bp window centered on the cleavage site to account for local sequence context and structural accessibility [116].
Table 2: Comparison of Major On-Target Efficiency Scoring Algorithms
| Algorithm Name | Basis of Development | Key Features Considered | Applications/Tools |
|---|---|---|---|
| Rule Set 1 [116] | Knock-out efficiency data of 1,841 sgRNAs | 30nt sequence including sgRNA binding area, PAM, and flanking sequences | CHOPCHOP |
| Rule Set 2 [116] | Data from 4,390 sgRNAs (expanded dataset) | Gradient-boosted regression trees; improved feature weighting | CHOPCHOP, CRISPOR |
| Rule Set 3 [116] | 7 existing gRNA efficiency datasets of 47k gRNAs | Incorporates tracrRNA sequence variations | GenScript, CRISPick |
| CRISPRscan [116] | Activity data of 1,280 gRNAs in zebra fish | In vivo validation; species-specific considerations | CHOPCHOP, CRISPOR |
| Lindel [116] | ~1.16 million mutation events from 6,872 targets | Predicts indel patterns and frameshift ratio using logistic regression | CRISPOR |
Diagram 1: Workflow for gRNA On-Target Efficiency Prediction
Strategies for Minimizing Off-Target Effects
Off-target effects represent a major concern in CRISPR applications, particularly for therapeutic development, as unintended edits can lead to detrimental consequences including genotoxicity [108] [120]. Off-target activity occurs when Cas9 cleaves genomic sites with significant homology to the intended target, particularly at sites with similar sequences but with mismatches, especially in the PAM-distal region [108].
Several algorithmic strategies have been developed to minimize off-target effects:
Comprehensive Homology Analysis: Early approaches focused on identifying sequences with high similarity to the gRNA across the genome. The general rule is that sequences with fewer than three nucleotide mismatches, particularly those with fewer mismatches in the PAM-proximal "seed" region, pose higher off-target risks [116]. Modern tools perform exhaustive genome-wide searches to enumerate all potential off-target sites.
Position-Specific Mismatch Scoring: Not all mismatches contribute equally to reducing off-target cleavage. Scoring systems like the Cutting Frequency Determination (CFD) score assign different weights to mismatches based on their position and nucleotide context [116]. Mismatches in the PAM-proximal region (positions 1-12) generally have greater impact in reducing off-target activity than those in the PAM-distal region.
Guide-Intrinsic Mismatch Tolerance (GMT): Recent research has revealed that some gRNAs exhibit intrinsic tolerance to mismatches, independent of the specific mismatch context [118]. This GMT effect correlates with sequence composition, with guanine-enriched and thymine-depleted protospacers showing higher mismatch tolerance. Algorithms now incorporate GMT predictions to select gRNAs with naturally higher specificity.
Advanced tools like GuideScan2 leverage innovative genome indexing approaches to comprehensively analyze gRNA specificity [120]. This tool uses a memory-efficient Burrows-Wheeler transform combined with simulated reverse-prefix trie traversals to identify all potential off-target sites, including those with non-canonical PAM sequences. Such comprehensive analysis is crucial, as studies have identified widespread confounding effects of low-specificity gRNAs in published CRISPR screens [120].
Table 3: Off-Target Prediction and Scoring Methods
| Method | Basis of Development | Scoring Approach | Applications |
|---|---|---|---|
| Homology Analysis [116] | Sequence similarity search | Counts mismatches between gRNA and off-target sites | Initial screening in multiple tools |
| MIT Specificity Score (Hsu Score) [116] | Indel mutation data from 700+ gRNA variants with 1-3 mismatches | Position-weighted mismatch scoring | Original MIT CRISPR design tool, CRISPOR |
| Cutting Frequency Determination (CFD) [116] | Activity data of 28,000 gRNAs with single variations | Matrix-based scoring of variations; multiplicative | GenScript, CRISPick |
| GuideScan2 Specificity Score [120] | Exhaustive genome indexing with mismatch and alternative PAM consideration | Specificity scoring based on off-target enumeration | GuideScan2 web and command-line tools |
Integrated gRNA Design Tools and Workflows
Several sophisticated software tools integrate both on-target efficiency and off-target specificity predictions to provide comprehensive gRNA design solutions. These tools implement the algorithms discussed previously and offer user-friendly interfaces for researchers [115] [116].
CRISPick (Broad Institute) incorporates Rule Set 3 for on-target scoring and CFD for off-target evaluation, providing a balanced approach to gRNA selection [116]. The tool is particularly valued for its rigorous benchmarking and regular updates incorporating the latest research findings.
GuideScan2 represents a significant advancement in gRNA design technology due to its novel genome indexing approach that enables memory-efficient, comprehensive off-target analysis [120]. Its ability to rapidly evaluate gRNA specificity across entire genomes has revealed previously unappreciated confounding effects in CRISPR screens, where genes targeted by low-specificity gRNAs were systematically under-called as hits in CRISPRi screens [120].
CHOPCHOP and CRISPOR are versatile tools that support multiple CRISPR systems beyond standard SpCas9 and provide visual representations of potential off-target sites [116]. These tools often incorporate multiple scoring algorithms side-by-side, allowing researchers to compare predictions and select the most reliable gRNAs.
When designing gRNAs for different application contexts, specific considerations apply:
- Gene Knockouts: Target early exons encoding critical protein domains, avoiding regions too close to N- or C-termini where truncated but functional proteins might be produced [115].
- Knock-ins: Prioritize gRNAs that cut close to the intended edit site, as HDR efficiency drops dramatically with increasing distance from the cleavage site [115].
- CRISPRi/a: Focus on target sites within promoter regions that are accessible for transcriptional regulation, balancing location constraints with sequence quality [115].
Diagram 2: gRNA Design and Selection Workflow
Experimental Validation and Analysis of Editing Outcomes
After computational design, experimental validation of gRNA performance remains essential. Several methods have been developed to assess both on-target efficiency and off-target activity [121].
For on-target validation, the gold standard is targeted next-generation sequencing (NGS), which provides comprehensive quantification of editing efficiency and characterization of the resulting indel spectrum [121]. However, NGS can be cost-prohibitive for large-scale screening. Alternative methods include:
- Inference of CRISPR Edits (ICE): A tool from Synthego that uses Sanger sequencing data to deconvolve complex editing outcomes and quantify efficiency [121]. ICE shows high correlation with NGS results (R² = 0.96) while being more accessible and cost-effective.
- TIDE (Tracking of Indels by Decomposition): An earlier decomposition method for Sanger sequencing data that provides estimation of indel frequencies but with more limited capability for complex edit patterns [121].
- T7 Endonuclease 1 (T7E1) Assay: A non-sequencing method that detects heteroduplex formation at edited sites but provides limited quantitative data and no information about specific indel sequences [121].
For off-target validation, several genome-wide methods have been developed:
- GUIDE-seq: Identifies off-target sites by capturing double-strand breaks with incorporated double-stranded oligonucleotides [108].
- BLESS: Direct in situ breaks labeling that can be applied to tissue samples without requiring exogenous bait molecules [108].
- Digenome-seq: An in vitro approach that sequences Cas9-digested genomic DNA to identify cleavage sites [108].
Recent advances in validation methodologies include the development of dual-target systems that measure relative cleavage rates between off-target and on-target sequences (off-on ratios) in a single assay. This approach provides internal normalization and reduces experimental variability, enabling more accurate quantification of specificity [118].
Table 4: The Scientist's Toolkit: Essential Reagents and Methods for gRNA Validation
| Reagent/Method | Function/Application | Key Features |
|---|---|---|
| Synthego ICE Analysis [121] | Computational analysis of Sanger sequencing data from edited cells | Cost-effective; provides indel spectrum and efficiency comparable to NGS |
| T7E1 Assay [121] | Detection of editing events via mismatch cleavage | Fast and inexpensive; no sequence information |
| Targeted NGS [121] | Deep sequencing of edited genomic regions | Gold standard; comprehensive but resource-intensive |
| GUIDE-seq [108] | Genome-wide identification of off-target sites | Comprehensive off-target profiling; requires dsODN delivery |
| Dual-Target System [118] | Measurement of relative off-target vs on-target cleavage | Internal normalization; reduced experimental variability |
Algorithmic gRNA design has evolved from simple sequence matching to sophisticated models that leverage large-scale experimental data and advanced machine learning techniques. The integration of both on-target efficiency and off-target specificity predictions has significantly improved the success rate of CRISPR experiments while reducing unintended consequences [116] [120].
Future directions in gRNA design include the development of cell-type-specific models that incorporate epigenetic features such as chromatin accessibility and DNA methylation patterns [114]. Additionally, as CRISPR therapeutics advance toward clinical applications, the demand for even higher specificity and the ability to design allele-specific gRNAs will increase [118] [113]. The recent discovery of "epistasis-like" combinatorial effects of multiple mismatches suggests new strategies for designing gRNAs that can discriminate between single nucleotide polymorphisms, enabling selective targeting of mutant alleles while sparing wild-type sequences [118].
The rapid pace of development in both computational algorithms and experimental validation methods ensures that gRNA design will continue to become more accurate and reliable. As these tools become more accessible and integrated into standardized workflows, researchers will be better equipped to harness the full potential of CRISPR technology for both basic research and therapeutic applications.
Validating CRISPR Systems: Analytical Methods, Clinical Evidence, and Technology Comparisons
Within the broader thesis on the basic principles of CRISPR-Cas9 genome editing research, the establishment of a robust analytical validation framework is paramount. The fundamental mechanism of CRISPR-Cas9 involves creating a double-strand break (DSB) in the DNA at a location specified by a guide RNA (gRNA), which is then repaired by the cell's endogenous repair pathways, primarily non-homologous end joining (NHEJ) or homology-directed repair (HDR) [16] [122]. Editing efficiency refers to the frequency at which these desired genetic alterations occur at the intended target site, while specificity denotes the system's ability to minimize unintended "off-target" edits at other genomic locations [123]. The accurate measurement of these two parameters is not merely a procedural step but a critical determinant of experimental reliability, reproducibility, and safety, especially in therapeutic contexts [124] [125]. This guide provides an in-depth technical framework for assessing these critical metrics, equipping researchers and drug development professionals with the protocols and analytical tools necessary to rigorously validate their CRISPR experiments.
Foundational Concepts: Editing Outcomes and Repair Pathways
A clear understanding of the molecular outcomes of CRISPR editing is a prerequisite for selecting appropriate validation strategies. When the Cas nuclease induces a DSB, the cell's repair machinery generates a spectrum of mutations.
- Non-Homologous End Joining (NHEJ): This is the dominant and error-prone repair pathway. It directly ligates the broken DNA ends, often resulting in small insertions or deletions (indels). When these indels occur within the coding region of a gene, they can disrupt the open reading frame, leading to a gene knockout [16] [122].
- Homology-Directed Repair (HDR): This pathway uses a donor DNA template with homologous sequences flanking the cut site to repair the break. While less efficient than NHEJ, HDR allows for precision editing, including the introduction of specific point mutations, gene insertions, or corrections [123] [122].
The following diagram illustrates the core workflow of a CRISPR-Cas9 experiment and the resulting repair outcomes that require validation.
A Toolkit for Validation: Methods for Assessing Editing Efficiency
Multiple experimental techniques are available to quantify the frequency of on-target edits, each with distinct strengths, sensitivities, and logistical considerations [126] [124]. The choice of method depends on the experimental goal, required sensitivity, throughput, and available resources.
Methodologies and Protocols
- T7 Endonuclease I (T7E1) Assay: This enzyme mismatch cleavage assay is a popular first-pass screening method due to its simplicity and speed [126].
- Protocol: Genomic DNA is harvested from the edited cell population and the target locus is amplified by PCR. A high-fidelity polymerase is critical to avoid PCR-introduced errors. The PCR products are denatured and reannealed, creating heteroduplexes where wild-type and mutant strands pair. These mismatched heteroduplexes are cleaved by the T7E1 enzyme. The products are separated by agarose gel electrophoresis, and the editing efficiency is estimated by comparing the band intensities of cleaved versus uncleaved products [126].
- Sanger Sequencing with Deconvolution Analysis: This method provides more detailed information than gel-based assays by directly examining the DNA sequence.
- Protocol: The target site is amplified by PCR from bulk genomic DNA (a heterogeneous mixture of edited and unedited cells) and subjected to Sanger sequencing. The resulting chromatogram, which shows overlapping signals downstream of the cut site, is analyzed by specialized software algorithms such as Tracking of Indels by DEcomposition (TIDE) or Inference of CRISPR Edits (ICE). These tools deconvolute the complex sequencing trace to quantify the spectrum and frequency of different indel mutations [126] [124].
- Targeted Amplicon Sequencing (Next-Generation Sequencing, NGS): This is considered the "gold standard" for comprehensive analysis due to its high sensitivity and accuracy [124].
- Protocol: The target region is PCR-amplified from genomic DNA with barcoded primers, enabling the pooling of hundreds of samples. The pooled library is sequenced on a high-throughput NGS platform. The resulting millions of reads are aligned to a reference sequence, allowing for the precise identification and quantification of every unique insertion, deletion, and substitution at the target site with very low detection limits (often <0.1%) [124] [125].
- Droplet Digital PCR (ddPCR): This method offers absolute quantification of specific edits without the need for a standard curve.
- Protocol: The technique involves partitioning the PCR reaction into thousands of nanoliter-sized droplets. Fluorescent probes are designed to distinguish between wild-type and mutant alleles. After endpoint PCR, the droplet reader counts the fraction of positive droplets for each fluorescence channel, enabling precise calculation of the allele frequency [124].
Comparative Analysis of Efficiency Assessment Methods
Table 1: A comparison of key methods for quantifying CRISPR editing efficiency.
| Method | Key Principle | Sensitivity | Throughput | Key Advantages | Key Limitations |
|---|---|---|---|---|---|
| T7E1 Assay [126] [124] | Enzyme cleavage of DNA heteroduplexes | Low-Moderate | Medium | Inexpensive, fast, no need for specialized software [126] | Does not identify specific sequence changes; can yield false positives [126] [124] |
| Sanger + TIDE/ICE [126] [124] | Deconvolution of bulk Sanger sequencing traces | Moderate (~5%) | Low-Medium | Cost-effective, provides indel sequence information [126] | Lower sensitivity than NGS; does not detect rare variants (<5%) well [124] |
| NGS (AmpSeq) [124] [125] | High-throughput sequencing of target amplicons | Very High (<0.1%) | High (when multiplexed) | Highly sensitive and accurate; provides full spectrum of edits [124] | Higher cost; longer turnaround time; complex data analysis [124] |
| ddPCR [124] | Absolute quantification via droplet partitioning | High (~0.1-1%) | Medium | High precision; no standard curve needed; excellent for validating specific edits [124] | Requires specific probe design; only detects pre-defined edits |
Ensuring Precision: Strategies for Evaluating Specificity and Off-Target Effects
A highly efficient editor is of little value if it also modifies unintended sites in the genome. A comprehensive validation framework must therefore include a rigorous assessment of specificity.
In Silico Prediction and Guide RNA Design
The first line of defense against off-target effects is careful gRNA design. Computational tools (e.g., CRISPOR) analyze the gRNA sequence for potential off-target sites across the genome based on sequence similarity, particularly in the "seed" region, and mismatch tolerance [124] [123]. Selecting gRNAs with minimal homology to other genomic regions is a critical first step. Furthermore, the use of high-fidelity Cas9 variants (e.g., eSpCas9, SpCas9-HF1, HypaCas9) engineered to reduce non-specific interactions with DNA can significantly lower off-target activity without compromising on-target efficiency [16].
Experimental Detection of Off-Target Edits
- Cell-Based Methods: Several methods leverage the cell's own repair machinery to report off-target activity. However, these can be limited by the specificity of the nuclease used in the assay itself.
- NGS-Based Profiling: These are the most comprehensive and unbiased methods.
- Circularization for In Vitro Reporting of Cleavage Effects by Sequencing (CIRCLE-Seq): This is a highly sensitive in vitro method. Genomic DNA is sheared, circularized, and then treated with the CRISPR RNP complex in vitro. Any linearized DNA fragments resulting from off-target cleavage are selectively amplified and sequenced via NGS, providing a genome-wide profile of potential cleavage sites [127].
- Single-Cell DNA Sequencing: An emerging powerful technology, single-cell DNA sequencing (e.g., Tapestri) allows for the simultaneous analysis of on-target and off-target edits across hundreds of loci in thousands of individual cells. This provides a unique view of editing outcomes, including the co-occurrence of edits (zygosity) and genomic instability within single cells, offering unprecedented insight for safety assessment [125].
The following workflow outlines a strategic approach to comprehensively assess both on- and off-target editing.
Successful execution of the validation framework relies on a suite of trusted reagents and tools. The following table catalogues key resources cited in the literature.
Table 2: Key research reagent solutions for CRISPR analytical validation.
| Reagent / Tool | Function / Description | Example Product / Source |
|---|---|---|
| High-Fidelity Cas9 | Engineered Cas9 variant with reduced off-target effects for higher specificity edits [16]. | eSpCas9(1.1), SpCas9-HF1 [16] |
| T7 Endonuclease I | Mismatch-specific endonuclease used in the T7E1 assay to detect heteroduplex DNA [126]. | Sigma-Aldrich T7E1 Kit [126] |
| Alt-R CRISPR-Cas9 System | Optimized synthetic CRISPR reagents (e.g., modified gRNAs) designed for improved efficiency and reduced toxicity [127]. | Integrated DNA Technologies (IDT) [127] |
| rhAmpSeq CRISPR Analysis System | A targeted amplicon sequencing system designed for sensitive and multiplexed on- and off-target analysis [127]. | Integrated DNA Technologies (IDT) [127] |
| CRISPR Plasmids | Pre-cloned, validated vectors for expressing Cas9, gRNAs, and base editors. | Addgene [16] [123] |
| Validation Controls | Pre-validated gRNAs and experimental baselines to ensure assay accuracy and instrument function [126]. | Positive control gRNAs (e.g., targeting housekeeping genes) [126] |
Advanced Topics: Enhancing Efficiency and Novel Detection Paradigms
The field of CRISPR analytics continues to evolve rapidly. Recent advances focus on both improving editing outcomes and deepening the analytical capabilities.
- Small Molecule Enhancers: Studies have identified small molecules that can modulate DNA repair pathways to favor desired outcomes. For instance, Repsox, a TGF-β signaling inhibitor, has been shown to promote NHEJ, increasing knockout efficiency by up to 3.16-fold in porcine cells [30]. Other molecules, like GSK-J4 and Zidovudine, have also demonstrated efficacy in enhancing NHEJ-mediated editing [30].
- Artificial Intelligence in Editor Design: Moving beyond naturally derived Cas enzymes, large language models (LMs) are now being used to design highly functional, novel genome editors. These AI-generated proteins, such as OpenCRISPR-1, can exhibit comparable or improved activity and specificity relative to SpCas9 while being vastly different in sequence, opening new avenues for optimized editing tools [46].
- The Rise of Single-Cell DNA Sequencing: As highlighted in a 2025 study, bulk sequencing methods can obscure the true complexity of editing outcomes in a heterogeneous cell population. Single-cell DNA sequencing enables the co-detection of on-target edits, off-target edits, and chromosomal rearrangements within the same cell, providing a new level of resolution for assessing the clonality and safety of edited cell products [125]. This is particularly crucial for clinical applications where genotoxicity is a major concern.
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9 system has revolutionized genome engineering by providing an unprecedented ability to perform precise, targeted modifications to the human genome. This RNA-guided system enables specific gene modulation through the Cas9 nuclease, which creates double-strand breaks (DSBs) at targeted genomic loci directed by a guide RNA (gRNA) sequence [84]. The cellular repair of these breaks through either non-homologous end joining (NHEJ) or homology-directed repair (HDR) pathways enables permanent genetic alterations with therapeutic intent. The transition of CRISPR-Cas9 from a research tool to clinical application represents a landmark achievement in genetic medicine, culminating in the first regulatory approvals of CRISPR-based therapies and a growing pipeline of investigational treatments across diverse disease areas [9] [84].
This technical guide comprehensively analyzes efficacy and safety data from approved CRISPR therapies and advanced clinical trials, framed within the fundamental principles of CRISPR-Cas9 genome editing research. We examine the specific molecular mechanisms, experimental methodologies, and clinical outcomes that demonstrate both the transformative potential and current limitations of this technology. The integration of quantitative clinical data with detailed experimental protocols provides researchers and drug development professionals with a rigorous framework for evaluating CRISPR-based therapeutic applications.
Approved CRISPR Therapies: Efficacy and Safety Profiles
CASGEVY (exagamglogene autotemcel, exa-cel)
CASGEVY, developed by CRISPR Therapeutics and Vertex Pharmaceuticals, represents the first FDA-approved CRISPR-based gene therapy, indicated for sickle cell disease (SCD) and transfusion-dependent beta thalassemia (TDT) [128]. This ex vivo therapy involves editing autologous CD34+ hematopoietic stem cells to disrupt the BCL11A gene, specifically in the erythroid lineage, thereby restoring fetal hemoglobin production.
Table 1: Clinical Efficacy Outcomes for CASGEVY in SCD and TDT
| Disease | Patient Population | Primary Endpoint | Efficacy Results | Duration of Follow-up |
|---|---|---|---|---|
| Sickle Cell Disease | Patients with history of vaso-occlusive crises (VOCs) | Freedom from severe VOCs | 94.5% of patients achieved freedom from severe VOCs | 24 months post-infusion |
| Transfusion-Dependent Beta Thalassemia | Patients requiring regular red blood cell transfusions | Transfusion independence | 91.7% of patients achieved transfusion independence | 24 months post-infusion |
The safety profile of CASGEVY is characterized primarily by the side effects associated with myeloablative conditioning with busulfan, which is required prior to infusion of the edited cells. Common adverse events include febrile neutropenia, stomatitis, and decreased appetite. To date, no evidence of off-target editing or insertional mutagenesis has been reported in clinical studies, though long-term follow-up continues as mandated by regulatory agencies [128].
CTX310: ANGPTL3-Targeted Therapy for Lipid Disorders
CTX310 represents a novel in vivo CRISPR-Cas9 therapeutic approach for managing dyslipidemias. This investigational therapy utilizes lipid nanoparticles (LNPs) to deliver CRISPR components targeting the angiopoietin-like 3 (ANGPTL3) gene in hepatocytes [129] [130]. ANGPTL3 regulates triglyceride and LDL metabolism, and natural loss-of-function mutations are associated with favorable lipid profiles and reduced cardiovascular risk.
Table 2: Efficacy Outcomes from Phase 1 Trial of CTX310 (0.8 mg/kg dose cohort)
| Parameter | Baseline Value | Mean Reduction | Maximum Reduction | Timepoint |
|---|---|---|---|---|
| Circulating ANGPTL3 | Variable | 73% | 89% | Day 30 |
| Triglycerides (TG) | >150 mg/dL | 55% (60% in elevated TG) | 84% | Day 60 |
| LDL Cholesterol | Variable | 49% | 87% | Day 60 |
In the Phase 1 clinical trial, CTX310 demonstrated a favorable safety profile with no treatment-related serious adverse events [129] [130]. Adverse events were generally mild to moderate, with three participants experiencing Grade 2 infusion-related reactions that resolved completely. One participant with elevated transaminases at baseline had a transient Grade 2 elevation that peaked by Day 4 and resolved completely by Day 14 without intervention. No ≥Grade 3 changes in liver transaminases were observed, supporting continued clinical development [129].
Experimental Protocols and Methodologies
CTX310 Phase 1 Trial Design
The Phase 1, open-label, dose-escalation trial evaluated single-course intravenous doses of CTX310 ranging from 0.1 to 0.8 mg/kg (lean body weight) in patients with various dyslipidemias [129] [130]. Key methodological elements included:
- Patient Populations: The trial enrolled 15 participants across four disease categories: homozygous familial hypercholesterolemia (HoFH), severe hypertriglyceridemia (sHTG), heterozygous familial hypercholesterolemia (HeFH), and mixed dyslipidemias.
- Inclusion Criteria: Eligible participants had uncontrolled TG levels >150 mg/dL and/or LDL cholesterol >100 mg/dL (or >70 mg/dL for those with established atherosclerotic cardiovascular disease) despite background standard of care per local guidelines.
- Background Therapy: The majority of participants were receiving statins and/or ezetimibe, while 40% were taking PCSK9 inhibitors, representing a treatment-resistant population.
- Endpoints: Primary endpoints focused on safety and tolerability, with secondary endpoints assessing changes in circulating ANGPTL3 protein, TG, and LDL levels.
- Premedication: All participants received corticosteroids and antihistamines prior to infusion to mitigate potential infusion-related reactions [130].
CASGEVY Clinical Trial Methodology
The global clinical development program for CASGEVY employed consistent methodology across multiple sites [128]:
- Cell Collection: CD34+ hematopoietic stem and progenitor cells were collected from patients via apheresis after mobilization with plerixafor.
- Ex Vivo Editing: Cells were edited using CRISPR-Cas9 to target a specific enhancer region in the first intron of the BCL11A gene, disrupting a GATA1 transcription factor binding site critical for BCL11A expression in erythroid cells.
- Manufacturing: The editing process occurred ex vivo under controlled manufacturing conditions without viral vectors, utilizing electroporation for delivery of CRISPR components.
- Conditioning and Infusion: Patients received myeloablative busulfan conditioning followed by infusion of the edited autologous CD34+ cells.
- Follow-up: Patients were monitored for engraftment, adverse events, and efficacy parameters with planned long-term follow-up of 15 years to assess durable safety.
Molecular Mechanisms and Signaling Pathways
The therapeutic mechanisms of approved CRISPR therapies illustrate sophisticated applications of fundamental genome editing principles. The following diagram illustrates the molecular mechanism of CTX310, which targets the ANGPTL3 gene in the liver:
Figure 1: Molecular mechanism of CTX310-mediated ANGPTL3 editing for lipid reduction.
The mechanism of CASGEVY operates through a different pathway, targeting the BCL11A gene to restore fetal hemoglobin production in red blood cells:
Figure 2: CASGEVY mechanism of action through BCL11A gene editing in hematopoietic stem cells.
Safety Considerations and Risk Mitigation
Off-Target Effects and Genomic Integrity
Comprehensive assessment of CRISPR-based therapies must include rigorous evaluation of potential off-target effects and structural variations. Recent studies have revealed that beyond simple insertions or deletions (indels), CRISPR-Cas9 editing can induce larger structural variations (SVs), including chromosomal translocations and megabase-scale deletions [27]. These findings highlight the importance of sensitive detection methods such as CAST-Seq and LAM-HTGTS to fully characterize the genomic landscape post-editing.
Notably, strategies to enhance homology-directed repair (HDR) through inhibition of key non-homologous end joining (NHEJ) pathway components like DNA-PKcs may inadvertently increase the frequency of these large-scale aberrations [27]. This demonstrates the complex balance between optimizing editing efficiency and maintaining genomic integrity, particularly in therapeutic contexts.
Long-Term Safety Monitoring
Regulatory agencies including the FDA and EMA require long-term safety monitoring for all CRISPR-based therapies, typically up to 15 years [130]. This monitoring includes:
- Regular assessment for potential late-onset adverse events
- Evaluation of clonal composition and dynamics in edited cell populations
- Monitoring for potential malignant transformation
- Assessment of durability of therapeutic effect
For in vivo therapies like CTX310, additional monitoring includes repeated assessment of liver function and broader metabolic parameters to detect any potential long-term consequences of hepatic gene editing [129].
The Scientist's Toolkit: Essential Research Reagents
Table 3: Key Research Reagents for CRISPR Clinical Development
| Reagent/Category | Function | Example Applications |
|---|---|---|
| Lipid Nanoparticles (LNPs) | In vivo delivery of CRISPR components | Hepatic gene editing (CTX310, CTX320) |
| Cas9 Nucleases | RNA-guided DNA cleavage | Gene knockout (ANGPTL3, BCL11A) |
| Guide RNA (gRNA) | Target sequence recognition | Specific gene targeting |
| AAV Vectors | In vivo delivery vehicle | Muscle tissue targeting (HG-302 for DMD) |
| CD34+ HSPCs | Target cells for ex vivo editing | Hematopoietic disorders (CASGEVY) |
| Homology-Directed Repair Templates | Precise gene correction | Gene knock-in strategies |
| Next-Generation Sequencing Assays | Off-target and structural variation analysis | CAST-Seq, LAM-HTGTS |
| Cell Sorting Technologies | Isolation and purification of edited cells | CD34+ selection for ex vivo therapies |
The clinical trial outcomes for approved CRISPR therapies demonstrate remarkable efficacy in treating genetic disorders, with manageable safety profiles in the studied populations. The durable responses observed with both ex vivo (CASGEVY) and in vivo (CTX310) approaches validate CRISPR-Cas9 genome editing as a transformative therapeutic modality. However, comprehensive long-term safety monitoring and continued refinement of editing precision remain essential as the field advances.
Future directions include the development of more precise editing tools such as base editors and prime editors, enhanced delivery systems for tissues beyond the liver, and approaches to minimize potential genotoxic risks [131] [84]. The integration of artificial intelligence into gRNA design and outcome prediction represents another promising frontier for optimizing therapeutic efficacy and safety [131]. As the clinical experience with CRISPR therapies expands, these data will inform the development of next-generation editors with improved precision and reduced off-target effects, ultimately broadening the therapeutic applications of this revolutionary technology.
The advent of engineered nucleases has revolutionized biological research and therapeutic development by enabling precise, targeted modifications to the genome. Three major technologies—Zinc Finger Nucleases (ZFNs), Transcription Activator-Like Effector Nucleases (TALENs), and the Clustered Regularly Interspaced Short Palindromic Repeats-associated system (CRISPR-Cas9)—comprise the core of this genome editing toolkit [132] [133]. Each system operates on a common principle: creating targeted double-strand breaks (DSBs) in DNA that stimulate the cell's innate repair mechanisms, primarily non-homologous end joining (NHEJ) or homology-directed repair (HDR) [132] [134]. While ZFNs and TALENs represent pioneering protein-based targeting systems, CRISPR-Cas9 has emerged as a more recent and transformative technology that utilizes RNA-DNA recognition [134]. This review provides a comprehensive technical comparison of these three genome editing platforms, examining their molecular mechanisms, experimental parameters, and relative advantages to inform selection for research and therapeutic applications.
Molecular Mechanisms and Design
Zinc Finger Nucleases (ZFNs)
ZFNs are fusion proteins comprising an array of engineered zinc finger proteins (ZFPs) fused to the FokI endonuclease domain [132] [133]. Each zinc finger domain recognizes a specific 3-4 base pair DNA sequence, with arrays typically containing 3-6 fingers to recognize 9-18 base pairs [133]. A critical feature of ZFNs is that the FokI nuclease domain must dimerize to become active, necessitating the design of two ZFN units that bind opposite DNA strands at sequences spaced 5-7 base pairs apart [134] [133]. This dimerization requirement enhances specificity but complicates design, as zinc finger motifs assembled in arrays can influence neighboring finger specificity through context-dependent effects [132] [134].
Transcription Activator-Like Effector Nucleases (TALENs)
TALENs similarly utilize the FokI nuclease domain but employ DNA-binding domains derived from transcription activator-like effectors (TALEs) [132] [133]. Each TALE repeat domain consists of 33-35 amino acids and recognizes a single DNA base pair through two hypervariable residues known as repeat-variable diresidues (RVDs) [132]. Specific RVD codes have been established: NI for adenine, NG for thymine, HD for cytosine, and NN for guanine/adenine [132]. Like ZFNs, TALENs function as pairs binding opposite DNA strands with intervening spacer sequences, and FokI dimerization is required for DNA cleavage [134]. The one-to-one correspondence between TALE repeats and nucleotides simplifies design compared to ZFNs, though cloning repetitive TALE arrays presents technical challenges [132].
CRISPR-Cas9 System
The CRISPR-Cas9 system fundamentally differs from ZFNs and TALENs by utilizing RNA-guided DNA recognition [134]. The system consists of the Cas9 nuclease and a single-guide RNA (sgRNA) approximately 100 nucleotides long that combines the functions of CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA) [50]. The sgRNA directs Cas9 to complementary DNA sequences adjacent to a protospacer adjacent motif (PAM), which for the commonly used Streptococcus pyogenes Cas9 (SpCas9) is 5'-NGG-3' [50] [134]. Cas9 induces a blunt-ended double-strand break approximately 3-4 nucleotides upstream of the PAM sequence [50]. DNA recognition occurs through Watson-Crick base pairing between the sgRNA and target DNA, simplifying redesign to new targets by modifying the sgRNA sequence alone [134].
Table 1: Fundamental Characteristics of Genome Editing Technologies
| Feature | ZFNs | TALENs | CRISPR-Cas9 |
|---|---|---|---|
| DNA Recognition Mechanism | Protein-DNA [134] | Protein-DNA [134] | RNA-DNA [134] |
| DNA Binding Domain | Zinc finger proteins (3-4 bp/finger) [133] | TALE repeats (1 bp/repeat) [132] | sgRNA (20 bp guide sequence) [50] |
| Nuclease Component | FokI endonuclease [133] | FokI endonuclease [134] | Cas9 endonuclease [50] |
| Dimerization Required | Yes [134] | Yes [134] | No [50] |
| Target Sequence Length | 9-18 bp per ZFN (18-36 bp total with spacer) [133] | 30-40 bp total (including spacer) [134] | 20 bp guide + PAM (5'-NGG-3') [50] [134] |
| PAM Requirement | None | None | Essential (5'-NGG-3' for SpCas9) [134] |
Performance Comparison and Experimental Considerations
Efficiency and Specificity
Direct comparative studies reveal significant differences in the efficiency and specificity of these genome editing platforms. A 2021 study using GUIDE-seq to evaluate off-target activity in HPV-targeted gene therapy found substantial variation in specificity [135]. ZFNs demonstrated variable but often massive off-target effects (287-1,856 off-target sites), with specificity reversibly correlated with the count of middle "G" in zinc finger proteins [135]. TALENs showed intermediate off-target activity, with design choices aimed at improving efficiency (such as αN-terminal domains or NN recognition modules) inevitably increasing off-target effects [135]. Notably, SpCas9 demonstrated superior specificity in this parallel comparison, with zero off-targets detected in URR and E6 regions and only 4 off-targets in the E7 region, compared to 36 off-targets for TALENs in E7 [135].
CRISPR-Cas9 generally offers higher editing efficiency in most experimental systems, though this comes with ongoing concerns about off-target effects [136]. Various strategies have been developed to enhance CRISPR-Cas9 specificity, including the use of high-fidelity Cas9 variants (HF-Cas9, eCas9, HypaCas9), Cas9 nickases that require paired recognition for double-strand breaks, and fusion of catalytically dead Cas9 (dCas9) with FokI nuclease [134]. For ZFNs and TALENs, off-target effects can be reduced through the use of obligate heterodimer FokI domains that prevent homodimerization [133].
Practical Implementation and Design Flexibility
From a practical standpoint, these technologies differ significantly in their design complexity, cloning requirements, and experimental flexibility:
ZFNs: Design is challenging due to context-dependent effects between neighboring zinc fingers [134]. Modular assembly requires pre-selected zinc finger libraries, and commercial sources may be necessary for reliable results [132] [133]. Target site density is limited, with effective targeting sites approximately every 200 bp using open-source components [133].
TALENs: More straightforward design based on the RVD code but complicated by highly repetitive sequences that make cloning technically challenging [132]. Golden Gate assembly and other specialized methods have been developed to facilitate TALEN construction [132]. TALENs are particularly advantageous for editing repetitive sequences or regions with high GC content where CRISPR-Cas9 may struggle [136].
CRISPR-Cas9: The simplest design system, requiring only the synthesis of a ~20 nucleotide guide RNA sequence complementary to the target DNA [136] [134]. This simplicity enables rapid targeting of multiple genomic sites simultaneously (multiplexing) and extensive genome-wide library screens [134]. The main constraint is the PAM requirement, though Cas9 variants with altered PAM specificities are expanding targeting range [134].
Table 2: Experimental Practicality and Applications
| Parameter | ZFNs | TALENs | CRISPR-Cas9 |
|---|---|---|---|
| Design Complexity | High (context-dependent effects) [134] [133] | Moderate (repetitive cloning challenges) [132] | Low (simple sgRNA design) [136] [134] |
| Cloning Process | Complex (engineering linkages between motifs) [134] | Moderate (Golden Gate assembly) [134] | Simple (sgRNA expression vector or direct RNA) [134] |
| Multiplexing Capacity | Limited | Limited | High (multiple sgRNAs) [134] |
| Targeting Density | ~1 site every 200 bp (open-source) [133] | High (theoretically any sequence) [132] | Limited by PAM frequency (~1/8-16 bp for NGG) [134] |
| Delivery Constraints | Protein coding sequence only | Large protein coding sequence | Cas9 protein/sgRNA or coding sequences |
Experimental Protocols and Workflows
Target Site Selection and Reagent Design
ZFN Design Protocol:
- Identify target sequence with form 5'-(NNN)₃-NNG-3' where NNG represents the preferred triplet for zinc finger binding [133]
- Select zinc finger modules from validated libraries using open-source platforms or commercial services [132] [133]
- Design ZFN pairs with 5-7 bp spacer between binding sites to allow FokI dimerization [133]
- Clone ZFN coding sequences into appropriate expression vectors
- Validate DNA binding affinity and specificity before nuclease testing [133]
TALEN Design Protocol:
- Identify target sequence with 5'-T-start requirement for natural TALEs [132]
- Design TALE repeat arrays using RVD code (NI-A, HD-C, NN-G, NG-T) [132]
- Design TALEN pairs with 14-20 bp spacer between binding sites [134]
- Assemble TALE repeats using Golden Gate cloning or similar methodology [132]
- Clone finalized TALEN arrays into expression vectors with FokI domain [132]
CRISPR-Cas9 Design Protocol:
- Identify target sequence with 5'-N₂₀-NGG-3' format immediately upstream of PAM [50]
- Design sgRNA with 20-nucleotide guide sequence complementary to target
- Check potential off-target sites using bioinformatics tools
- Clone sgRNA sequence into expression vector with U6 promoter or synthesize as RNA [134]
- Select appropriate Cas9 variant (wild-type, nickase, high-fidelity) for application [134]
Delivery Methods and Validation
Delivery considerations vary significantly between platforms and experimental systems:
Physical Methods: Microinjection (common for embryonic editing), hydrodynamic injection (preclinical models), electroporation (effective for hard-to-transfect cells including immune cells and stem cells) [50]
Carrier-Based Methods: Lipid nanoparticles (increasingly used for CRISPR component delivery, particularly for liver targets), extracellular vesicles (natural carriers with biocompatibility advantages) [50]
Viral Vectors: Adeno-associated viruses (AAVs, limited packaging capacity of ~4.7 kb challenging for SpCas9 at ~4.2 kb), lentiviral vectors (larger capacity, integration concerns) [50]
Each delivery method presents tradeoffs between efficiency, cytotoxicity, and persistence that must be optimized for specific applications. Recent advances have demonstrated successful in vivo delivery of CRISPR-Cas9 via lipid nanoparticles for therapeutic applications including hereditary transthyretin amyloidosis (hATTR) and hereditary angioedema (HAE) [9].
Diagram 1: Genome editing workflow showing key experimental stages and delivery methods.
Research Reagent Solutions
Successful implementation of genome editing technologies requires appropriate selection of core reagents and tools. The following table outlines essential research reagents and their functions:
Table 3: Essential Research Reagents for Genome Editing
| Reagent Category | Specific Examples | Function | Technology Application |
|---|---|---|---|
| Nuclease Components | ZFP arrays, TALE repeats, Cas9 variants | DNA recognition and cleavage | All platforms |
| Expression Vectors | CMV-driven Cas9, U6-driven sgRNA | Component expression in target cells | All platforms, particularly CRISPR |
| Delivery Tools | Electroporation systems, Lipid nanoparticles (LNPs), AAV vectors | Efficient component delivery to cells | All platforms |
| Validation Reagents | T7E1 assay, GUIDE-seq, NGS libraries | Detection and quantification of editing | All platforms |
| Repair Templates | ssODNs, dsDNA donors with homology arms | HDR-mediated precise editing | All platforms |
| Cell Culture Supplements | Chemical enhancers (e.g., Rad51 stimulators), Antibiotic selection | Improve HDR efficiency, select edited cells | All platforms |
Current Applications and Future Perspectives
The genome editing field has expanded dramatically from basic research to therapeutic applications. CRISPR-Cas9 has dominated recent clinical translation, with the first FDA-approved CRISPR therapy (Casgevy for sickle cell disease and beta thalassemia) marking a milestone [9]. Ongoing clinical trials demonstrate expanding applications, including Intellia Therapeutics' phase III trial for hereditary transthyretin amyloidosis using LNP-delivered CRISPR-Cas9 [9]. The personalized CRISPR treatment developed for an infant with CPS1 deficiency further demonstrates the technology's potential, with development and FDA approval achieved in just six months [9].
The global market landscape reflects these technological shifts, with the genome editing market projected to grow from $10.8 billion in 2025 to $23.7 billion by 2030, representing a 16.9% compound annual growth rate [137] [138]. CRISPR technology dominates this market expansion, though TALENs and ZFNs maintain specific niches where their protein-based targeting offers advantages [137].
Future directions include the development of more precise editing tools such as base editors and prime editors, enhanced delivery systems particularly for non-liver targets, and continued refinement of specificity through high-fidelity variants and improved bioinformatic prediction of off-target effects [50] [9]. The integration of artificial intelligence and machine learning is poised to further enhance the accuracy and efficiency of all genome editing platforms [50].
Diagram 2: Technology evolution and current clinical status of genome editing platforms.
The comparative analysis of ZFNs, TALENs, and CRISPR-Cas9 reveals a complex landscape where each technology offers distinct advantages for specific applications. ZFNs provide high specificity when properly designed but present significant technical challenges. TALENs offer simplified design and robust activity across diverse genomic contexts but involve cloning difficulties. CRISPR-Cas9 delivers unprecedented simplicity, flexibility, and multiplexing capacity, though PAM requirements and off-target concerns remain considerations.
For most research applications, CRISPR-Cas9 represents the most practical and efficient choice, particularly when rapid testing of multiple targets is required. However, TALENs maintain value for challenging targets where CRISPR efficiency is low or when protein-based recognition is preferred. ZFNs, while historically important, have largely been superseded except in specialized applications where extensive optimization has already been performed.
As the field advances, the convergence of these technologies—such as FokI-dCas9 fusions that combine RNA-guided targeting with FokI dimerization—may further blur distinctions between platforms. The fundamental understanding of all three systems remains essential for researchers to select appropriate tools and interpret results within the expanding genome editing toolkit.
The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) systems represent a revolutionary genome engineering technology derived from adaptive immune mechanisms in prokaryotes [20] [16]. These systems recognize and cleave foreign genetic elements through programmable RNA-guided complexes, providing unprecedented precision in manipulating DNA and RNA across diverse applications from basic research to clinical therapeutics [94] [20]. The core principle involves two fundamental components: a guide RNA (gRNA) that specifies the target sequence through complementary base pairing, and a Cas nuclease that executes cleavage of the target nucleic acid [94] [16].
CRISPR-Cas systems are broadly categorized into two classes based on their effector complex architecture. Class 1 systems (types I, III, and IV) utilize multi-subunit protein complexes for target interference, while Class 2 systems (types II, V, and VI) employ single effector proteins, making them particularly suitable for genome engineering applications [139] [40]. This technical guide focuses on three predominant Class 2 CRISPR systems: Cas9 (type II), Cas12 (type V), and Cas13 (type VI), which have become indispensable tools in modern molecular biology and therapeutic development [140] [40].
Comparative Analysis of Cas Enzyme Families
Structural Characteristics and Mechanisms
Cas9: The DNA Targeting Workhorse The Cas9 nuclease, particularly from Streptococcus pyogenes (SpCas9), functions as a multi-domain DNA endonuclease requiring both a CRISPR RNA (crRNA) and trans-activating CRISPR RNA (tracrRNA), which can be fused into a single-guide RNA (sgRNA) [20] [139]. Structurally, Cas9 exhibits a bilobed architecture consisting of a recognition (REC) lobe and a nuclease (NUC) lobe [139]. The REC lobe (REC1-3 domains) facilitates sgRNA binding and wrapping, while the NUC lobe contains the RuvC and HNH nuclease domains along with a Protospacer Adjacent Motif (PAM)-interacting (PI) domain [20] [139]. The HNH domain cleaves the DNA strand complementary to the sgRNA (target strand), while the RuvC domain cleaves the non-complementary strand (non-target strand), resulting in a blunt-ended double-strand break (DSB) approximately 3-4 nucleotides upstream of the PAM sequence [16] [139]. Cas9 activation involves precise conformational checkpoints, including PAM recognition, seed sequence annealing, R-loop formation, and final nuclease domain activation [139].
Cas12: The Staggered DNA Cutter Cas12 (formerly Cpf1) represents a distinct family of type V CRISPR-Cas systems with several structural and functional differences from Cas9 [140]. Unlike Cas9, Cas12 proteins require only a crRNA for guidance and lack the tracrRNA component [94] [140]. Cas12 enzymes recognize T-rich PAM sequences (typically TTN or TTTN) and generate staggered DNA breaks with 5' overhangs rather than blunt ends [140]. The Cas12 family includes multiple variants such as Cas12a, Cas12b, and Cas12f, with Cas12a being the most extensively characterized [94] [141]. Structurally, Cas12 contains a single RuvC nuclease domain responsible for cleaving both DNA strands [139]. A distinctive feature of Cas12 enzymes is their collateral cleavage activity (trans-cleavage), where upon target recognition, they non-specifically degrade single-stranded DNA [142] [140]. This property has been harnessed for diagnostic applications.
Cas13: The RNA-Targeting Specialist Cas13 represents a family of type VI CRISPR-Cas systems that exclusively target RNA rather than DNA [140]. Like Cas12, Cas13 requires only a crRNA for guidance and possesses collateral RNAse activity upon target recognition [142] [140]. Cas13 enzymes contain two Higher Eukaryotes and Prokaryotes Nucleotide-binding (HEPN) domains that mediate RNA cleavage [139]. Upon binding to its target RNA, Cas13 undergoes a conformational change that activates its HEPN domains, leading to cleavage of the target RNA and subsequent non-specific collateral trans-cleavage of nearby non-target RNA molecules [142] [140]. This RNA-targeting capability enables transient modulation of gene expression without permanent genomic changes, making Cas13 particularly valuable for diagnostic applications, RNA knockdown, and potential antiviral therapeutic strategies [142] [140].
Quantitative Comparison of Key Properties
Table 1: Structural and Functional Characteristics of Major Cas Enzymes
| Property | Cas9 | Cas12a | Cas13a |
|---|---|---|---|
| Class/Type | Class 2, Type II | Class 2, Type V | Class 2, Type VI |
| Target Nucleic Acid | dsDNA | dsDNA/ssDNA | ssRNA |
| gRNA Composition | crRNA + tracrRNA (or sgRNA) | crRNA only | crRNA only |
| gRNA Size | ~100 nt | ~40 nt | ~50 nt |
| PAM Sequence | 3'-NGG-5' (SpCas9) | 5'-TTTN-3' | Protospacer Flanking Site (PFS): A, U, or C (LshCas13a) |
| Cleavage Products | Blunt ends | Staggered ends (5' overhangs) | RNA fragments |
| cis-Cleavage | dsDNA | dsDNA/ssDNA | ssRNA |
| trans-Cleavage | No | ssDNA | ssRNA |
| Size (aa) | 1368 (SpCas9) | 1200-1300 (Cas12a) | ~1150 (LshCas13a) |
| Nuclease Domains | HNH, RuvC | RuvC | 2× HEPN |
Table 2: Performance Metrics and Applications of Cas Enzymes
| Parameter | Cas9 | Cas12 | Cas13 |
|---|---|---|---|
| Editing Efficiency | High (varies by target) | High (varies by variant) | High for RNA knockdown |
| Off-Target Effects | Moderate to High | Moderate | Moderate to High (due to collateral activity) |
| Multiplexing Capacity | Moderate (requires additional gRNAs) | High (self-processing pre-crRNA) | High (self-processing pre-crRNA) |
| Primary Applications | Gene knockout, Gene insertion, Genome-wide screens | Gene editing, DNA detection (DETECTR) | RNA knockdown, RNA editing, Viral RNA detection (SHERLOCK) |
| Therapeutic Delivery | Challenging due to large size | More feasible (smaller variants available) | Feasible (moderate size) |
| Clinical Stage | Approved (Casgevy for SCD/TBT) | Phase I/II trials (e.g., HG302 for DMD) | Preclinical (diagnostics in development) |
Experimental Protocols for Nuclease Characterization
Protocol 1: Assessing Genome Editing Efficiency
Objective: Quantify the targeted editing efficiency of Cas nucleases in mammalian cells.
Materials and Reagents:
- Plasmid vectors encoding Cas nuclease (e.g., pCas9, pCas12a, pCas13a)
- Guide RNA expression vectors or synthetic gRNAs
- Mammalian cell line (HEK293T commonly used)
- Transfection reagent (e.g., Lipofectamine 3000)
- Lysis buffer for genomic DNA extraction
- PCR purification kit
- T7 Endonuclease I or TIDE analysis reagents
- Next-generation sequencing library preparation kit
Methodology:
- Design and Cloning: Design gRNAs targeting specific genomic loci of interest. For Cas9, ensure presence of 5'-NGG-3' PAM adjacent to target; for Cas12a, identify 5'-TTTN-3' PAM. Clone gRNA sequences into appropriate expression vectors [141] [16].
- Cell Transfection: Culture HEK293T cells in appropriate medium. Co-transfect cells with Cas nuclease expression plasmid and gRNA plasmid at optimal ratio (typically 1:1 to 1:3 ratio). Include negative controls (cells transfected with Cas plasmid only) [141].
- Genomic DNA Extraction: Harvest cells 72 hours post-transfection. Extract genomic DNA using standard phenol-chloroform method or commercial kits.
- Editing Efficiency Analysis:
- T7E1 Assay: Amplify target region by PCR (30-35 cycles). Denature and reanneal PCR products in thermocycler. Digest heteroduplex DNA with T7 Endonuclease I for 30 minutes. Analyze fragments by agarose gel electrophoresis. Calculate editing efficiency based on band intensities [141].
- Next-Generation Sequencing: Amplify target regions with barcoded primers. Prepare sequencing libraries and sequence on Illumina platform. Analyze sequencing data using CRISPResso2 or similar tools to quantify indel frequencies [141].
Data Interpretation: Editing efficiency is calculated as percentage of modified alleles in the cell population. Cas9 typically achieves 30-60% efficiency in human cell lines, while Cas12 variants may show comparable or slightly reduced efficiency depending on target site [141].
Protocol 2: Evaluating Off-Target Effects
Objective: Identify and quantify off-target editing activities of Cas nucleases.
Materials and Reagents:
- Cas-gRNA ribonucleoprotein (RNP) complexes
- Genomic DNA from treated cells
- Whole genome amplification kit
- Hybridization capture reagents
- Next-generation sequencing platform
- Bioinformatics analysis tools (Cas-OFFinder, GUIDE-seq)
Methodology:
- In Silico Prediction: Use computational tools to predict potential off-target sites with sequence similarity to the intended target, allowing up to 5 nucleotide mismatches [94] [16].
- GUIDE-seq (Genome-wide Unbiased Identification of DSBs Enabled by Sequencing):
- Transfect cells with Cas-gRNA complex along with GUIDE-seq oligonucleotides.
- Allow 72 hours for integration of oligonucleotides at DSB sites.
- Extract genomic DNA and prepare sequencing libraries.
- Enrich for integrated oligonucleotides and sequence.
- Map double-strand break sites throughout the genome [16].
- Targeted Deep Sequencing: Design primers for predicted off-target sites. Amplify regions of interest and perform deep sequencing (≥10,000x coverage). Quantify indel frequencies at each potential off-target site [94].
Data Interpretation: Compare indel frequencies at off-target sites to background mutation rates. High-fidelity variants (e.g., eSpCas9, SpCas9-HF1) should show significantly reduced off-target editing (<0.1% at most sites) compared to wild-type nucleases [94] [16].
Protocol 3: Bacterial Plasid Eradication Assay
Objective: Assess the efficiency of different CRISPR systems in eliminating antibiotic resistance genes from bacterial plasmids.
Materials and Reagents:
- Recombinant CRISPR plasmids (pCas9, pCas12f1, pCas3)
- E. coli strains carrying KPC-2 or IMP-4 carbapenem resistance plasmids
- LB broth and agar plates
- Antibiotics: tetracycline, chloramphenicol, gentamicin, kanamycin
- Restriction enzymes (BsaI)
- Rapid ligation kit
- Colony PCR reagents
- Drug sensitivity test materials
Methodology:
- Target Design and Plasmid Construction: Design spacers within specific regions of resistance genes (e.g., 542-576 bp of KPC-2 gene). For Cas9, select 30-nt sequence upstream of NGG PAM; for Cas12f1, 20-nt upstream of TTTN; for Cas3, 34-nt antisense strand upstream of GAA motif [141].
- Plasmid Assembly: Digest CRISPR plasmids with BsaI restriction enzyme. Ligate with annealed spacer oligonucleotides using rapid ligation kit. Transform into competent E. coli cells [141].
- Eradication Efficiency Assessment: Introduce recombinant CRISPR plasmids into model drug-resistant bacteria. Plate transformations on selective media. Perform colony PCR to confirm eradication of resistance genes. Conduct drug sensitivity tests to verify resensitization to antibiotics [141].
- Quantitative PCR: Extract plasmids from transformants. Perform qPCR to measure copy number changes of drug-resistant plasmids. Use comparative Ct method to calculate eradication efficiency [141].
Data Interpretation: CRISPR-Cas3 systems have demonstrated higher eradication efficiency (up to 100% for KPC-2 and IMP-4 genes) compared to Cas9 and Cas12f1 in quantitative assessments [141].
Visualization of CRISPR Mechanisms
Diagram 1: Comparative Mechanisms of Cas9, Cas12, and Cas13 Nucleases. The diagram illustrates the distinct activation pathways and cleavage activities of the three major Cas enzyme families, highlighting key differences in PAM requirements, cleavage patterns, and collateral activities.
Research Reagent Solutions
Table 3: Essential Research Reagents for CRISPR-Cas Experiments
| Reagent Category | Specific Examples | Function & Application | Key Considerations |
|---|---|---|---|
| Cas Expression Plasmids | pSpCas9(BB) (Addgene #48138), pCas12a (Cpf1), pC013 (Cas13a) | Delivery of nuclease component; stable or transient expression | Choose backbone with appropriate promoters for target cells; consider size constraints for viral packaging [141] [16] |
| Guide RNA Vectors | pU6-gRNA, pMBP-gRNA, multiplex gRNA arrays | Expression of target-specific guide RNAs | U6 promoter works well in mammalian cells; multiplex vectors enable simultaneous targeting of multiple loci [16] |
| CRISPR Delivery Systems | Lentiviral vectors, AAV vectors, Lipid Nanoparticles (LNPs) | Efficient intracellular delivery of CRISPR components | AAV has limited cargo capacity (~4.7kb); LNPs suitable for RNP delivery; consider immunogenicity for in vivo applications [94] [9] |
| Validation Tools | T7E1 assay reagents, GUIDE-seq oligos, NGS library prep kits | Assessment of editing efficiency and specificity | T7E1 provides rapid screening; NGS offers comprehensive off-target profiling; GUIDE-seq identifies genome-wide DSBs [141] [16] |
| Cell Culture Reagents | Transfection reagents (lipofectamine, PEI), selection antibiotics (puromycin, blasticidin) | Introduction and maintenance of CRISPR components in cells | Optimization required for different cell types; primary cells often require RNP delivery for high efficiency [141] |
| Efficiency Enhancers | HDR enhancers (RS-1, L755507), nuclear localization signals | Improve editing outcomes and precision | Particularly important for HDR-mediated precise edits; NLS tags critical for nuclear import in non-dividing cells [20] |
Applications in Research and Therapeutics
Therapeutic Genome Editing
CRISPR-Cas systems have demonstrated remarkable potential in treating genetic disorders through precise genome manipulation. The first FDA-approved CRISPR therapy, Casgevy, utilizes Cas9 for treating sickle cell disease and transfusion-dependent beta thalassemia by editing the BCL11A gene to restore fetal hemoglobin production [9] [80]. Clinical trials have expanded to include various genetic disorders, with over 250 gene-editing clinical trials currently monitored, spanning blood disorders, cancers, metabolic diseases, and infectious diseases [9] [80].
Notable advances include Intellia Therapeutics' phase I trial for hereditary transthyretin amyloidosis (hATTR) using Cas9 delivered via lipid nanoparticles (LNPs), representing the first systemic in vivo CRISPR-Cas9 therapy [9]. Results demonstrated sustained ~90% reduction in disease-related TTR protein levels with minimal side effects, highlighting the potential of LNP delivery for targeting the liver [9]. Similarly, CRISPR-based approaches are being investigated for hereditary angioedema (HAE), with phase I/II trials showing 86% reduction in kallikrein protein and significant reduction in disease attacks [9].
Diagnostic Applications
The collateral cleavage activities of Cas12 and Cas13 have been harnessed for developing highly sensitive diagnostic platforms. Cas12 targets DNA and exhibits collateral ssDNA cleavage, while Cas13 targets RNA with collateral ssRNA cleavage, enabling detection of specific nucleic acid sequences with attomolar sensitivity when coupled with isothermal amplification methods [142].
The Specific High Sensitivity Enzymatic Reporter UnLOCKing (SHERLOCK) platform utilizes Cas13 for detecting RNA viruses, including SARS-CoV-2 lineage differentiation, while DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR) employs Cas12 for DNA target detection [142]. These systems provide rapid, cost-effective diagnostic solutions suitable for point-of-care testing, with operational simplicity and the ability to integrate specific nucleic acid sequence detection without complex instrumentation [142]. Recent advances focus on achieving single-nucleotide specificity through strategic gRNA design, effector selection, and optimized reaction conditions [142].
Agricultural and Biotechnology Applications
CRISPR-Cas systems have been widely adopted for crop improvement, with SaCas9 used to edit genomes in tobacco, potato, and rice to enhance stress tolerance and pathogen resistance [94]. Studies comparing editing efficiency in plants found SaCas9 most efficient at generating indels, highlighting the importance of nuclease selection for specific applications [94]. Beyond crop improvement, CRISPR systems show promise in industrial biotechnology for metabolic engineering of production strains and developing novel biomaterials [20].
Emerging Trends and Future Perspectives
The CRISPR-Cas field continues to evolve with several emerging trends shaping future research directions. The discovery and characterization of novel Cas variants, such as the recently identified type VII systems containing Cas14 effectors, expands the available toolkit for specialized applications [40]. Additionally, engineering efforts continue to enhance existing nucleases, with developments like high-fidelity Cas12Max variants demonstrating improved editing efficiency with reduced off-target effects [94].
Delivery challenges remain a significant focus, with advances in viral and non-viral delivery systems critical for therapeutic applications. The successful use of lipid nanoparticles for in vivo delivery represents a significant breakthrough, enabling redosing possibilities not feasible with viral vectors due to immune responses [9]. The landmark case of a personalized in vivo CRISPR therapy developed for an infant with CPS1 deficiency in just six months demonstrates the potential for rapid development of bespoke treatments for rare genetic disorders [9].
Computational approaches and artificial intelligence are increasingly being leveraged for gRNA design and outcome prediction, addressing challenges in specificity and efficiency [142]. As the CRISPR toolbox expands to include base editors, prime editors, and epigenetic modifiers, the precision and scope of genome engineering applications continue to grow, promising new therapeutic modalities for addressing previously untreatable genetic diseases [94] [80].
The therapeutic potential of any genome editing intervention is ultimately determined by the durability of the intended genomic alteration and the persistence of its functional effect. For CRISPR-Cas9-based therapies, long-term follow-up (LTFU) is therefore a critical component of both preclinical and clinical development, providing essential insights into the stability of edits, potential for delayed adverse events, and overall therapeutic sustainability [143] [144]. This technical guide examines the core principles, methodologies, and emerging data surrounding the durability of CRISPR-mediated genome editing, with a specific focus on implications for basic research and therapeutic development.
The fundamental goal of LTFU is to determine whether a single editing intervention can produce a lasting, potentially lifelong therapeutic benefit. This requires careful assessment of both the persistence of the edited genomic state in the originally modified cells and the maintenance of the edited cell population over time, particularly in self-renewing tissues [23]. The durability of editing outcomes is influenced by a complex interplay of factors including the targeted cell type, the specific editing approach employed, the efficiency of the initial editing event, and the selective advantage or disadvantage conferred by the edit.
Quantitative Evidence of Editing Durability from Clinical Trials
Recent clinical trials have yielded the first robust, long-term human data on the persistence of CRISPR-mediated edits. The following table summarizes key quantitative findings from prominent clinical trials that have reported durability outcomes over extended periods.
Table 1: Long-Term Durability Data from Select Clinical Trials of CRISPR-Based Therapies
| Therapy / Trial | Target / Condition | Delivery Method | Follow-Up Period | Key Durability Findings | Reference |
|---|---|---|---|---|---|
| Casgevy (exa-cel) | BCL11A / Sickle Cell Disease & β-Thalassemia | Ex Vivo (Electroporation) | >24 months | Stable fetal hemoglobin levels; persistent editing of hematopoietic stem/progenitor cells leading to functional cure. | [9] [145] |
| NTLA-2001 (Intellia) | TTR / hATTR Amyloidosis | In Vivo (LNP) | 24 months | ~90% reduction in serum TTR protein sustained in all 27 participants who reached 2-year follow-up. | [9] [146] |
| NTLA-2002 (Intellia) | KLKB1 / Hereditary Angioedema (HAE) | In Vivo (LNP) | 16 weeks (reported) | 86% reduction in kallikrein sustained; 8 of 11 high-dose participants were attack-free. | [9] |
| BRL-201 (Non-viral CAR-T) | PDCD1 / Lymphoma | Ex Vivo (Non-viral) | >60 months | Documented remission for over 5 years in an initial patient, indicating persistence of the edited CAR-T cell population. | [147] |
These data demonstrate that CRISPR-mediated edits can be highly durable, with effects persisting for multiple years in both ex vivo and in vivo applications. The sustained reduction of target proteins in the systemic in vivo trials (e.g., NTLA-2001) is particularly significant, as it indicates that the edited hepatocytes, which are long-lived, are maintaining the edited genotype and phenotype without apparent silencing or loss [9].
Methodological Framework for Assessing Durability
Establishing a robust LTFU protocol requires a multi-faceted approach that assesses editing at the genomic, functional, and cellular levels. The workflow below outlines the key components of a comprehensive durability assessment strategy.
Diagram 1: LTFU Experimental Workflow. This workflow outlines the integration of temporal sampling with multi-modal analysis to comprehensively assess editing durability.
Core Analytical Techniques
The following experimental protocols are fundamental for generating high-quality LTFU data.
Protocol 1: Longitudinal Tracking of Editing Efficiency and Outcomes
- Objective: To quantitatively measure the percentage of edited alleles and characterize the spectrum of indel mutations over time.
- Procedure:
- Sample Collection: Isolate genomic DNA from target cells or tissue at predetermined time points (e.g., 1 week, 1, 3, 6, 12, 18, 24 months post-intervention).
- PCR Amplification: Amplify the on-target genomic region using high-fidelity DNA polymerase. Include barcodes to mitigate PCR amplification bias.
- Next-Generation Sequencing (NGS): Perform deep amplicon sequencing (recommended coverage >100,000x) to detect low-frequency edits.
- Bioinformatic Analysis: Process raw sequencing data using specialized pipelines (e.g., CRISPResso2, guideseq) to calculate:
- Editing Efficiency: (Total edited reads / Total aligned reads) * 100.
- Indel Spectrum: The distribution and frequency of specific insertion and deletion mutations.
- Allele Heterogeneity: The diversity of edited sequences present in the population [23].
Protocol 2: Assessment of Functional Persistence
- Objective: To confirm that the genomic edit translates into a stable, functional consequence, such as sustained protein knockdown or transgene expression.
- Procedure:
- Protein Quantification:
- For knock-down strategies (e.g., hATTR, HAE): Use ELISA or mass spectrometry to quantify target protein levels in serum (TTR, kallikrein) at regular intervals [9].
- For knock-in strategies: Use flow cytometry or immunohistochemistry to detect the presence of the newly expressed protein (e.g., CAR on T cells, fetal hemoglobin in erythrocytes).
- Phenotypic Assessment:
- Protein Quantification:
Mechanisms Influencing Editing Persistence
The long-term stability of a CRISPR-induced edit is governed by several biological mechanisms, which differ significantly between dividing and non-dividing cells.
Cellular Division and Edit Dilution
In self-renewing tissues (e.g., hematopoietic system), the durability of an edit depends on the successful engraftment and persistence of edited long-term repopulating hematopoietic stem cells (LT-HSCs). An edit will only be permanent if it is harbored within this specific, self-renewing progenitor population. If only short-term progenitors are edited, the therapeutic effect will be transient, lasting only for the lifespan of the differentiated cell types [143].
DNA Repair in Non-Dividing Cells
Many therapeutic targets are in non-dividing (post-mitotic) cells, such as neurons and cardiomyocytes. Recent research reveals that the DNA repair landscape in these cells is fundamentally different from that in dividing cells, which directly impacts the timeline and outcome of editing.
- Slower Repair Kinetics: Cas9-induced double-strand breaks (DSBs) in neurons and cardiomyocytes are resolved over a much longer period (up to two weeks) compared to dividing cells, where repair typically plateaus within days. This suggests a more protracted and potentially different repair process in post-mitotic cells [23].
- Divergent Repair Pathways: Dividing cells like iPSCs preferentially use repair pathways like microhomology-mediated end joining (MMEJ), which often results in larger deletions. In contrast, neurons predominantly use non-homologous end joining (NHEJ), leading to a different spectrum of indel outcomes, typically characterized by a higher ratio of insertions to deletions and smaller indels [23].
The diagram below illustrates the critical mechanistic differences in how dividing and non-dividing cells process and maintain CRISPR-induced edits over the long term.
Diagram 2: Durability Mechanisms in Dividing vs. Non-Dividing Cells. The diagram contrasts the DNA repair pathways and long-term fate of edits in different cell types, which is a primary determinant of durability.
The Scientist's Toolkit: Essential Reagents and Solutions
Successful LTFU studies rely on a suite of specialized reagents and tools. The following table details key solutions for critical steps in the durability assessment workflow.
Table 2: Essential Research Reagents for Durability and Persistence Studies
| Reagent / Tool Category | Specific Example | Function in LTFU Studies |
|---|---|---|
| High-Fidelity DNA Polymerase | Q5 Hot Start (NEB), KAPA HiFi | Accurate amplification of the target locus from genomic DNA for NGS, minimizing PCR-induced errors that could be mistaken for rare edits. |
| NGS Amplicon Library Prep Kits | Illumina DNA Prep, Swift Accel | Preparation of sequencing-ready libraries from PCR amplicons for deep sequencing to quantify editing efficiency and indel spectra. |
| Bioinformatics Pipelines | CRISPResso2, guideseq | Automated, standardized analysis of NGS data to calculate editing percentages, map indel distributions, and assess allele heterogeneity. |
| Protein Quantification Assays | ELISA Kits, SIMOA, MSD | Highly sensitive quantification of target protein levels in serum or cell lysates to correlate genomic edits with functional persistence. |
| HDR Enhancer | Alt-R HDR Enhancer Protein (IDT) | Boosts homology-directed repair efficiency in hard-to-edit primary cells (e.g., iPSCs, HSCs), which is crucial for achieving lasting knock-in edits. |
| Virus-Like Particles (VLPs) | VSVG/BRL-co-pseudotyped FMLV VLPs | Efficient delivery of Cas9-RNP to difficult-to-transfect non-dividing cells (e.g., neurons, cardiomyocytes) for in vivo durability modeling. |
The collective evidence from preclinical models and clinical trials strongly indicates that CRISPR-Cas9 genome editing can yield stable, long-lasting effects, with documented persistence now extending beyond five years in some cases. The durability of an edit is not a single property but an emergent property of the interaction between the editing strategy, the target cell's biology, and the DNA repair machinery. For dividing tissues, engagement of long-term repopulating stem cells is paramount. For non-dividing tissues, the unique and slower repair mechanisms present both a challenge for initial efficiency and an opportunity for stable, lifelong correction once achieved.
Future directions in the field will focus on further refining LTFU methodologies, understanding the long-term biological impacts of editing in diverse cell types, and developing next-generation editing tools (e.g., base and prime editors) that may offer even greater stability and safety profiles. The ongoing collection and analysis of long-term data remain the cornerstone for realizing the full potential of CRISPR-based therapies as durable and potentially curative treatments for human disease.
The advent of CRISPR-Cas9 genome editing has catalyzed a new era in therapeutic development, enabling precise genetic modifications that were previously unimaginable. As these innovative treatments transition from laboratory research to clinical applications, navigating the evolving regulatory pathways has become paramount for researchers and drug development professionals. Regulatory agencies worldwide, including the U.S. Food and Drug Administration (FDA) and the European Medicines Agency (EMA), have developed specialized frameworks to address the unique challenges posed by CRISPR-based therapies [148] [149]. These frameworks balance the need for rigorous safety assessment with the flexibility required to accelerate transformative treatments for patients with serious diseases.
Understanding these regulatory pathways is essential for designing robust preclinical and clinical development plans. The regulatory landscape encompasses guidance on product characterization, preclinical testing, clinical trial design, and long-term follow-up, all tailored to the specific attributes of genome editing products [149]. This guide provides a comprehensive overview of current regulatory considerations, highlighting recent developments that are shaping the approval pathways for CRISPR-based therapeutics within the broader context of basic CRISPR-Cas9 research principles.
Regulatory Framework and Guidance Documents
FDA Regulatory Structure and Guidance
The FDA's Center for Biologics Evaluation and Research (CBER) oversees the regulation of CRISPR-based therapies through its Office of Therapeutic Products (OTP), a "super office" established to enhance review capabilities for advanced therapies [148]. This office comprises six specialized divisions covering gene therapy chemistry, manufacturing, and controls (CMC); cellular therapy and human tissue CMC; plasma protein therapeutics CMC; clinical evaluation; pharmacology/toxicology; and review management and regulatory review. To address the increasing complexity of these products, the FDA has recruited extensively, with the OTP reaching 75-80% staffing as of early 2024 [148].
The FDA has issued numerous guidance documents specifically addressing cell and gene therapy products. Key finalized guidances relevant to CRISPR therapeutics include "Human Gene Therapy Products Incorporating Human Genome Editing" (January 2024), "Considerations for the Development of Chimeric Antigen Receptor (CAR) T Cell Products" (January 2024), and "Studying Multiple Versions of a Cellular or Gene Therapy Product in an Early-Phase Clinical Trial" (November 2022) [149]. These documents provide recommendations on investigational new drug (IND) application requirements, manufacturing considerations, and clinical trial design.
Recent draft guidances reflect the FDA's adaptive approach to emerging technologies. These include "Expedited Programs for Regenerative Medicine Therapies for Serious Conditions" (September 2025), "Innovative Designs for Clinical Trials of Cellular and Gene Therapy Products in Small Populations" (September 2025), and "Frequently Asked Questions — Developing Potential Cellular and Gene Therapy Products" (November 2024) [149]. These documents demonstrate the agency's commitment to addressing the unique challenges of gene therapy development, particularly for rare diseases with limited patient populations.
Advanced Regulatory Pathways and Novel Approaches
Expedited Programs and Bespoke Therapy Pathways
The FDA has established specialized pathways to accelerate the development of promising therapies. The "Expedited Programs for Regenerative Medicine Therapies for Serious Conditions" provides guidance on the Regenerative Medicine Advanced Therapy (RMAT) designation, which CRISPR Therapeutics obtained for CTX112 in relapsed or refractory follicular lymphoma and marginal zone lymphoma [150] [149]. This designation facilitates more efficient development through intensive FDA-sponsor interactions.
A groundbreaking development is the FDA's "plausible mechanism" pathway, announced in November 2025 [151]. Designed for bespoke therapies for ultra-rare diseases, this pathway addresses cases where traditional clinical trials are not feasible due to极小 patient populations. To qualify, treatments must target the known biological cause of a disease, with developers providing well-characterized historical data on disease natural history [151]. Companies must also confirm through biopsy or preclinical tests that the treatment successfully edits its target and improves outcomes.
This pathway was inspired by cases like Baby KJ, an infant with a unique mutation causing CPS1 deficiency who received a personalized CRISPR treatment developed, FDA-approved, and delivered within six months [9] [151] [152]. The FDA will initiate an approval process for developers that meet objectives in "several consecutive patients with different bespoke therapies," after which companies must accumulate evidence showing continued benefit without serious harm [151].
Umbrella Trial Designs and Master Protocols
The FDA's guidance "Studying Multiple Versions of a Cellular or Gene Therapy Product in an Early-Phase Clinical Trial" outlines the agency's approach to umbrella trials, which employ a master protocol to evaluate multiple therapy versions simultaneously [148]. This design is particularly valuable for CRISPR therapies, where different delivery vectors or editing approaches may need comparison.
Table: FDA IND Requirements for Umbrella Trials of CRISPR Therapies
| Component | Primary IND A | Secondary INDs (B, C, D...) |
|---|---|---|
| Master Protocol | Included in full | Cross-referenced to Primary IND A |
| CMC Information | For product version A | For respective product versions B, C, D... |
| Pharmacology/Toxicology Data | For product version A | For respective product versions B, C, D... |
| Administrative | Primary application | Cross-references master protocol |
The guidance clarifies that different versions of a CRISPR therapy (e.g., AAV vectors with different capsid proteins) constitute distinct products requiring separate INDs, even when tested under the same master protocol [148]. This approach accelerates development by enabling direct comparison of different versions with a shared control group, reducing the total number of subjects needed—a significant advantage for rare disease research where patient recruitment is challenging.
Preclinical and Clinical Development Considerations
Preclinical Safety Assessment and Editing Validation
Robust preclinical assessment is fundamental to CRISPR therapeutic development. Regulatory guidances emphasize comprehensive evaluation of on-target editing efficiency, off-target effects, and long-term safety. The "Preclinical Assessment of Investigational Cellular and Gene Therapy Products" guidance outlines requirements for pharmacology and toxicology studies, including assessments of biodistribution, tumorigenicity, and immunogenicity [149].
A critical consideration is the detection of structural variations (SVs) beyond simple insertions or deletions (indels). Recent research has revealed that CRISPR editing can induce large SVs, including chromosomal translocations and megabase-scale deletions, particularly when DNA-PKcs inhibitors are used to enhance homology-directed repair (HDR) [27]. Traditional short-read sequencing often misses these large alterations because they delete primer-binding sites, leading to overestimation of precise editing rates [27]. Regulatory agencies now recommend specialized methods like CAST-Seq and LAM-HTGTS to detect these SVs [27].
Table: Methods for Assessing CRISPR Editing Outcomes in Preclinical Studies
| Assessment Type | Standard Methods | Advanced Methods | Regulatory Considerations |
|---|---|---|---|
| On-Target Editing | Sanger sequencing, Short-read NGS | Long-read sequencing (Nanopore, PacBio) | Quantification of HDR vs. NHEJ outcomes |
| Off-Target Effects | In silico prediction, GUIDE-seq | CIRCLE-seq, DISCOVER-Seq | Assessment of predicted and unpredited sites |
| Structural Variations | Karyotyping, FISH | CAST-Seq, LAM-HTGTS | Detection of large deletions, translocations |
| Functional Effects | Cell viability assays, Transcriptomics | Tumorigenicity studies, Immune response profiling | Evaluation of oncogenic potential, immunogenicity |
For in vivo therapies, biodistribution studies must track editing components to target and non-target tissues, assessing potential germline editing risks. The guidance "Long Term Follow-up After Administration of Human Gene Therapy Products" recommends 15 years of monitoring for patients receiving gene therapies to evaluate long-term risks [149].
Clinical Trial Design and Safety Monitoring
Clinical development of CRISPR therapies follows a phased approach, with early-phase trials focusing on safety and dosage determination, and later phases evaluating efficacy [9]. Adaptive trial designs are particularly valuable given the novel mechanisms of action and often small patient populations.
The FDA's "Innovative Designs for Clinical Trials of Cellular and Gene Therapy Products in Small Populations" draft guidance (September 2025) provides recommendations for statistical approaches and endpoint selection when traditional randomized controlled trials are not feasible [149]. These include Bayesian methods, historical controls, and composite endpoints that may be particularly relevant for CRISPR therapies targeting rare diseases.
Safety monitoring must address CRISPR-specific concerns, including immune responses to editing components (Cas protein, delivery vectors) and potential genotoxicity from off-target editing or structural variations [50] [27]. The FDA requires comprehensive long-term follow-up plans—15 years for integrating vectors and 5 years for non-integrating vectors—to monitor for delayed adverse events [149].
Manufacturing and Quality Control Requirements
Chemistry, Manufacturing, and Controls (CMC)
The "Chemistry, Manufacturing, and Control (CMC) Information for Human Gene Therapy Investigational New Drug Applications (INDs)" guidance outlines requirements for manufacturing process characterization, validation, and control [149]. For CRISPR therapies, this includes comprehensive documentation of editing components (Cas protein, guide RNA), delivery systems (viral vectors, LNPs), and final product characterization.
A significant challenge in CRISPR manufacturing is maintaining consistency in editing efficiency and product purity. The "Potency Assurance for Cellular and Gene Therapy Products" draft guidance (December 2023) emphasizes the need for quantitative potency assays that directly measure biological activity [149]. For CRISPR-based products, this may include direct measurement of target sequence modification, functional assays demonstrating intended physiological effects, and characterization of editing heterogeneity.
Table: Essential Research Reagent Solutions for CRISPR Therapeutic Development
| Reagent Category | Specific Examples | Function in Development | Regulatory Considerations |
|---|---|---|---|
| Editing Enzymes | High-fidelity Cas9, HiFi Cas9 variants [27] | Target DNA cleavage with reduced off-target effects | GMP-grade requirement for clinical use |
| Delivery Systems | AAV vectors, Lipid Nanoparticles (LNPs) [9] [50] | In vivo delivery of editing components | Purity, potency, and characterization data |
| Guide RNA | Modified sgRNA with enhanced stability [50] | Target sequence recognition | Verification of sequence fidelity and purity |
| DNA Repair Templates | Single-stranded DNA, AAV HDR templates [27] | Facilitate precise genetic corrections | Sequence verification, absence of contaminants |
| Analytical Tools | CAST-Seq reagents, NGS libraries [27] | Detection of on/off-target editing and SVs | Validation of sensitivity and specificity |
Delivery System Characterization and Testing
Delivery systems for CRISPR components require extensive characterization. For viral vectors like AAVs, this includes assessments of capsid purity, vector potency, and vector genome integrity [50]. For non-viral delivery systems like lipid nanoparticles (LNPs), critical quality attributes include particle size distribution, encapsulation efficiency, and stability [9] [150]. The "Recommendations for Microbial Vectors Used for Gene Therapy" guidance provides specific recommendations for viral vector testing [149].
The "Manufacturing Changes and Comparability for Human Cellular and Gene Therapy Products" draft guidance (July 2023) addresses the challenge of manufacturing process changes, which are common as therapies progress from research to commercial scale [149]. Sponsors must demonstrate that process modifications do not adversely affect critical quality attributes, including editing efficiency, product purity, and safety profile.
Emerging Challenges and Future Directions
Safety Considerations and Risk Mitigation
As CRISPR therapies advance clinically, understanding and mitigating potential risks remains paramount. Beyond the well-documented concern of off-target effects at sites with sequence similarity to the target, recent evidence highlights the significance of on-target structural variations, including chromosomal translocations and megabase-scale deletions [27]. These SVs raise substantial safety concerns, particularly for ex vivo edited therapies like Casgevy, where kilobase-scale deletions at the BCL11A target locus in hematopoietic stem cells have been documented [27].
Risk mitigation strategies include the use of high-fidelity Cas variants (e.g., HiFi Cas9) and careful selection of target sites to minimize off-target activity [27]. However, these approaches do not eliminate the risk of on-target SVs. The field is developing more sophisticated analytical methods to detect these alterations, and regulatory agencies increasingly expect comprehensive assessments of genomic integrity [27].
Another emerging challenge is the balance between editing efficiency and safety. Strategies to enhance HDR efficiency, such as using DNA-PKcs inhibitors, have been shown to exacerbate genomic aberrations, including dramatic increases in chromosomal translocations [27]. This highlights the need for careful evaluation of editing enhancement approaches and consideration of whether maximal editing efficiency is always necessary, particularly when corrected cells may have a selective advantage [27].
Regulatory Evolution and International Harmonization
The regulatory landscape for CRISPR therapies continues to evolve rapidly. The FDA's new "plausible mechanism" pathway for bespoke therapies represents a significant innovation in regulatory science, potentially enabling treatment for patients with ultra-rare genetic mutations [151] [152]. However, challenges in scaling these approaches remain, as exemplified by the resource-intensive development of Baby KJ's personalized therapy [152].
Internationally, regulatory harmonization efforts are ongoing. The EMA has developed scientific guidelines on gene therapy to help developers prepare marketing authorization applications [153]. While specific guidelines for CRISPR-based products are still emerging, the general principles for gene therapy evaluation apply, with particular emphasis on comprehensive risk-benefit assessment for innovative therapies.
As the field matures, regulatory agencies are increasing their capacity and expertise. The FDA's OTP continues to expand its staffing, and the START (Support for clinical Trials Advancing Rare disease Therapeutics) program enhances communication between sponsors and regulators for rare disease therapies [148]. These developments signal a growing recognition of the unique challenges and opportunities presented by CRISPR-based therapeutics.
CRISPR Therapy Development Pathways
CRISPR Mechanism and Editing Outcomes
Conclusion
CRISPR-Cas9 genome editing has evolved from a fundamental bacterial immune mechanism to a sophisticated therapeutic platform with demonstrated clinical success, as evidenced by approved treatments for sickle cell disease and beta-thalassemia. The integration of AI and machine learning is revolutionizing gRNA design and specificity prediction, while next-generation editors like base and prime editors are expanding the therapeutic landscape beyond double-strand breaks. Despite significant progress, challenges remain in delivery efficiency, tissue-specific targeting, and long-term safety monitoring. Future directions will focus on developing more precise delivery systems, expanding the scope of editable genetic targets, and establishing streamlined regulatory pathways for personalized CRISPR therapies. As clinical evidence accumulates and technology advances, CRISPR-based genome editing is poised to transform treatment paradigms across a broad spectrum of genetic disorders, cancers, and infectious diseases, ultimately realizing the promise of precision genetic medicine.
References
- 1. CRISPR/Cas9 therapeutics: progress and prospects [https://www.nature.com/articles/s41392-023-01309-7]
- 2. CRISPR-Cas9: From a bacterial immune system to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5470512/]
- 3. A Brief History of CRISPR-Cas9 Genome-Editing Tools [https://bitesizebio.com/47927/history-crispr/]
- 4. CRISPR Timeline [https://www.broadinstitute.org/what-broad/areas-focus/project-spotlight/crispr-timeline]
- 5. Evolution of CRISPR/Cas Systems for Precise Genome ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC10532350/]
- 6. CRISPR/Cas9 technology in tumor research and drug ... [https://www.frontiersin.org/journals/pharmacology/articles/10.3389/fphar.2025.1552741/full]
- 7. Cornerstones of CRISPR-Cas in drug development and therapy [https://pmc.ncbi.nlm.nih.gov/articles/PMC5459481/]
- 8. Principles of CRISPR-Cas9 technology: Advancements in ... [https://www.sciencedirect.com/science/article/pii/S1773224724000066]
- 9. CRISPR Clinical Trials: A 2025 Update [https://innovativegenomics.org/news/crispr-clinical-trials-2025/]
- 10. Scientists develop technology that brings new precision to ... [https://www.broadinstitute.org/news/technology-brings-new-precision-to-genome-editing]
- 11. CRISPR-Cas9 and Its Bioinformatics Tools: A Systematic ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12109910/]
- 12. Importance of the PAM Sequence in CRISPR Experiments [https://www.synthego.com/guide/how-to-use-crispr/pam-sequence/]
- 13. PAM identification by CRISPR-Cas effector complexes [https://pmc.ncbi.nlm.nih.gov/articles/PMC6546366/]
- 14. What Is the PAM Sequence for CRISPR and Where Is It? | IDT [https://www.idtdna.com/pages/support/faqs/what-is-a-pam-sequence-and-where-is-it-located]
- 15. Insights into the compact CRISPR–Cas9d system [https://www.nature.com/articles/s41467-025-57455-9]
- 16. CRISPR Guide [https://www.addgene.org/guides/crispr/]
- 17. Visualization of a multi-turnover Cas9 after product release [https://www.nature.com/articles/s41467-025-60668-7]
- 18. Quantification of the affinities of CRISPR–Cas9 nucleases ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7212624/]
- 19. A boost for the precision of genome | MIT News editing [https://news.mit.edu/2025/boost-precision-genome-editing-0820]
- 20. and Applications of Mechanism / CRISPR - Cas -Mediated 9 ... Genome [https://pmc.ncbi.nlm.nih.gov/articles/PMC8388126/]
- 21. DNA repair pathway choices in CRISPR-Cas9 mediated ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8187289/]
- 22. Visualizing the Nucleome Using the CRISPR–Cas9 System [https://pmc.ncbi.nlm.nih.gov/articles/PMC9940691/]
- 23. Characterizing and controlling CRISPR repair outcomes in ... [https://www.nature.com/articles/s41467-025-66058-3]
- 24. Uncovering the dynamics of precise repair at CRISPR ... [https://www.nature.com/articles/s41467-024-49410-x]
- 25. HDR vs. NHEJ: CRISPR Gene Editing Techniques [https://www.zeclinics.com/blog/hdr-vs-nhej-in-crispr-gene]
- 26. CRISPR-Cas9-mediated homology-directed repair for ... [https://www.sciencedirect.com/science/article/pii/S2162253124002312]
- 27. The hidden risks of CRISPR/Cas: structural variations and ... [https://www.nature.com/articles/s41467-025-62606-z]
- 28. Comparative analysis of multiple DNA double-strand break ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12075812/]
- 29. Weaponizing CRISPR/Cas9 for selective elimination of ... [https://www.sciencedirect.com/science/article/pii/S1568786425000369]
- 30. Improving the Efficiency of CRISPR/Cas9-Mediated Non ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12384319/]
- 31. Single-stranded DNA (ssDNA) donor repair templates and ... [https://www.frontiersin.org/journals/genome-editing/articles/10.3389/fgeed.2025.1661829/full]
- 32. Protocol to screen chemicals to enhance homology ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12491152/]
- 33. CRISPR-Cas systems: Overview, innovations and applications ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7508700/]
- 34. crispr [https://en.wikipedia.org/wiki/CRISPR]
- 35. Harnessing CRISPR-Cas system diversity for gene editing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8038530/]
- 36. Diversity and evolution of class 2 CRISPR–Cas systems [https://pmc.ncbi.nlm.nih.gov/articles/PMC5851899/]
- 37. Classy CRISPR: Differences between Class 1 and Class 2 ... [https://www.synthego.com/blog/crispr-systems/]
- 38. Discovery and functional characterization of diverse Class ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4660269/]
- 39. Typing CRISPR Systems [https://blog.addgene.org/typing-crispr-systems]
- 40. An updated evolutionary classification of CRISPR–Cas ... [https://www.nature.com/articles/s41564-025-02180-8]
- 41. Recent Updates of the CRISPR/Cas9 Genome Editing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11164216/]
- 42. Press release: The Nobel Prize in Chemistry 2020 [https://www.nobelprize.org/prizes/chemistry/2020/press-release/]
- 43. Nobel Prize Awarded to Jennifer Doudna And Emmanuelle ... [https://www.synthego.com/blog/gene-editing-nobel-prize/]
- 44. Basic Principles and Clinical Applications of CRISPR ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8819410/]
- 45. Recent advances in therapeutic gene-editing technologies [https://www.sciencedirect.com/science/article/pii/S152500162500200X]
- 46. Design of highly functional genome editors by modelling ... [https://www.nature.com/articles/s41586-025-09298-z]
- 47. CRISPR Technologies for In Vivo and Ex Vivo Gene Editing [https://www.ncbi.nlm.nih.gov/books/NBK609557/]
- 48. What are genome editing and CRISPR-Cas9? [https://medlineplus.gov/genetics/understanding/genomicresearch/genomeediting/]
- 49. innovations and challenges in rAAV vector-based CRISPR ... [https://www.nature.com/articles/s41434-025-00573-2]
- 50. Challenges and Opportunities in the Application of CRISPR ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12503735/]
- 51. CRISPR Cell and Gene Therapies: Key Challenges in ... [https://www.synthego.com/blog/crispr-cell-gene-therapy-obstacles/]
- 52. CRISPR Therapeutics Highlights Strategic Priorities and ... [https://ir.crisprtx.com/news-releases/news-release-details/crispr-therapeutics-highlights-strategic-priorities-and]
- 53. New Gene-Editing Strategy Could Help Development ... [https://www.nytimes.com/2025/11/19/health/gene-editing-rare-diseases.html]
- 54. Overcoming the AAV Size Limitation for CRISPR Delivery [https://blog.addgene.org/a-match-made-in-heaven-crispr/cas9-and-aav]
- 55. Adeno-associated virus-mediated delivery of CRISPR-Cas9 ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8153090/]
- 56. AAV-based CRISPR-Cas9 genome editing: Challenges ... [https://www.sciencedirect.com/science/article/pii/S2468451123000739]
- 57. AAV vs Lentivirus – Which Viral Vector Is Right For My ... [https://bioinnovatise.com/articles/aav-vs-lentivirus-which-viral-vector-is-right-for-my-research/]
- 58. Lentiviral Vectors for Delivery of Gene-Editing Systems Based ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC8310029/]
- 59. AAV Vs. Lentiviral Vectors - Life in the Lab [https://www.thermofisher.com/blog/life-in-the-lab/aav-vs-lv/]
- 60. CRISPR-Cas9 Lentivirus [https://www.thermofisher.com/us/en/home/life-science/genome-editing/cas9-nucleases/cas9-lentivirus.html]
- 61. Lentivirus CRISPR Vector [https://en.vectorbuilder.com/resources/vector-system/pLV_gRNA_Cas9_parent.html]
- 62. Comparative analysis of lipid Nanoparticle-Mediated ... [https://www.sciencedirect.com/science/article/pii/S093964112400033X]
- 63. Delivery Strategies of the CRISPR-Cas9 Gene-Editing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5723556/]
- 64. CRISPR-Cas9 Gene Therapy: Non-Viral Delivery and ... [https://www.mdpi.com/1420-3049/30/3/542]
- 65. Overcoming the Delivery Challenges in CRISPR/Cas9 ... [https://www.medsci.org/v22p3625.htm]
- 66. CRISPR Delivery Systems for Organ-Specific Targeting [https://www.sciencedirect.com/science/article/pii/S2950482125000336]
- 67. CRISPR/Cas9 Delivery Systems to Enhance Gene Editing ... [https://www.mdpi.com/1422-0067/26/9/4420]
- 68. Lipid nanoparticles outperform electroporation in mRNA- ... [https://www.sciencedirect.com/science/article/pii/S232905012300178X]
- 69. Comparative Analysis of CRISPR/Cas9 Delivery Methods ... [https://www.mdpi.com/1422-0067/26/21/10703]
- 70. Protocol for in vitro transfection of mRNA-encapsulated ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12597280/]
- 71. cutting-edge applications of base editing and prime editing in ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC11662623/]
- 72. Emerging trends in prime editing for precision genome ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12322275/]
- 73. Back to Basics - Base & Prime Editing [https://frontlinegenomics.com/back-to-basics-base-and-prime-editing/]
- 74. Explainer: What Is Prime Editing and What Is It Used For? [https://crisprmedicinenews.com/news/explainer-what-is-prime-editing-and-what-is-it-used-for/]
- 75. Prime editing: therapeutic advances and mechanistic insights [https://www.nature.com/articles/s41434-024-00499-1]
- 76. CRISPR-Cas9: a prominent genome editing tool in the ... [https://www.sciencedirect.com/science/article/abs/pii/S2452318625000406]
- 77. CRISPR/Cas technologies in pancreatic cancer research ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12339858/]
- 78. CRISPR-Cas9: a prominent genome editing tool in the ... [https://pubmed.ncbi.nlm.nih.gov/40753800/]
- 79. Panorama of CRISPR Gene Editing Clinical Applications [https://kactusbio.com/blogs/gene-editing/panorama-of-crispr-gene-editing-clinical-applications-2024-review?srsltid=AfmBOopqTMXWAxjrByTMvzes1Wc8yzQXHHgNzvTk7lC_Adan9z51mUPI]
- 80. Overview CRISPR Clinical Trials 2025 - Learn | Innovate [https://crisprmedicinenews.com/clinical-trials/]
- 81. The promise of CRISPR/Cas9 technology in diabetes ... [https://www.sciencedirect.com/science/article/abs/pii/S1056872723001228]
- 82. The next act for mRNA: pushing boundaries in cell and ... [https://www.pharmaceutical-technology.com/sponsored/pushing-boundaries-in-cell-and-gene-therapy/]
- 83. CRISPR Clinical Trials to Follow [https://www.synthego.com/blog/crispr-clinical-trials/]
- 84. Advancing CRISPR genome editing into gene therapy clinical ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12094669/]
- 85. CRISPR Therapies Clinical Trial Pipeline Shows Potential [https://www.globenewswire.com/news-release/2025/10/02/3160607/0/en/CRISPR-Therapies-Clinical-Trial-Pipeline-Shows-Potential-with-Active-Contributions-from-25-Key-Companies-DelveInsight.html]
- 86. The CRISPR therapeutics landscape in 2025 [https://www.kilburnstrode.com/knowledge/european-ip/the-crispr-therapeutics-landscape-in-2025]
- 87. Press Release [https://ir.crisprtx.com/news-releases/news-release-details/crispr-therapeutics-reports-positive-additional-phase-1-data]
- 88. Off-target effects in CRISPR/Cas9 gene editing - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC10034092/]
- 89. Exploring CRISPR-Cas: The transformative impact of gene ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12506487/]
- 90. Methods for detecting off-target effects of CRISPR/Cas9 [https://pubmed.ncbi.nlm.nih.gov/41213415/]
- 91. CRISPR Off-Target Editing: Prediction, Analysis, and More [https://www.synthego.com/blog/crispr-off-target-editing/]
- 92. Methods for detecting off-target effects of CRISPR/Cas9 [https://www.sciencedirect.com/science/article/abs/pii/S0734975025002368]
- 93. Deep learning predicts CRISPR off-target effects [https://crisprmedicinenews.com/news/deep-learning-predicts-crispr-off-target-effects/]
- 94. How to Choose the Right Cas Variant for Every CRISPR ... [https://www.synthego.com/guide/how-to-use-crispr/cas-nuclease-variants/]
- 95. Recent Advances in Improving Gene-Editing Specificity ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9319960/]
- 96. High-fidelity CRISPR-Cas9 variants with undetectable ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4851738/]
- 97. Enhanced proofreading governs CRISPR-Cas9 targeting ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC5918688/]
- 98. Structural insights into a high fidelity variant of SpCas9 [https://www.nature.com/articles/s41422-018-0131-6]
- 99. SpCas9-HF1 enhances accuracy of cell cycle-dependent ... [https://www.sciencedirect.com/science/article/pii/S2162253124000118]
- 100. Can SpRY recognize any PAM in human cells? - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC9110322/]
- 101. PAM-flexible genome editing with an engineered chimeric ... [https://www.nature.com/articles/s41467-023-41829-y]
- 102. Flexibility in PAM recognition expands DNA targeting ... [https://elifesciences.org/articles/102538]
- 103. qEva-CRISPR: a method for quantitative evaluation of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC6158505/]
- 104. CRISPRainbow and Genome Visualization [https://blog.addgene.org/crisprainbow-and-genome-visualization]
- 105. CRISPR Delivery Methods: Cargo, Vehicles, and Challenges [https://www.synthego.com/blog/delivery-crispr-cas9/]
- 106. Optimized design parameters for CRISPR Cas9 and ... [https://www.nature.com/articles/s41598-021-98965-y]
- 107. Design Rules for Expanding PAM Compatibility in CRISPR ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12424760/]
- 108. Methods for Optimizing CRISPR-Cas9 Genome Editing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC4976696/]
- 109. CRISPR/Cas9 gene editing: a novel strategy for fighting drug ... [https://biosignaling.biomedcentral.com/articles/10.1186/s12964-024-01713-8]
- 110. Peeling back the layers of immunogenicity in Cas9-based ... [https://www.sciencedirect.com/science/article/pii/S1525001625005350]
- 111. Non-clinical safety considerations on genome editing using ... [https://www.sciencedirect.com/science/article/pii/S2352304225002740]
- 112. Scientists engineer CRISPR enzymes that evade ... [https://www.broadinstitute.org/news/scientists-engineer-crispr-enzymes-evade-immune-system]
- 113. / CRISPR therapeutics: progress and prospects Cas 9 [https://www.nature.com/articles/s41392-023-01309-7?error=cookies_not_supported]
- 114. CRISPR–Cas9 gRNA efficiency prediction: an overview of ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC9023298/]
- 115. How To Design Guide RNA for CRISPR [https://www.synthego.com/guide/how-to-use-crispr/design-grna-crispr/]
- 116. A Guide to Efficient CRISPR gRNA Design: Principles and ... [https://www.genscript.com/a-guide-to-efficient-crispr-grna-design-principles-and-design-tools.html]
- 117. Rapid two-step target capture ensures efficient - CRISPR - Cas ... 9 guided [https://pubmed.ncbi.nlm.nih.gov/40273916/]
- 118. Systematic decomposition of sequence determinants ... [https://www.nature.com/articles/s41467-022-28028-x]
- 119. Sequence determinants of improved CRISPR sgRNA design [https://pmc.ncbi.nlm.nih.gov/articles/PMC4509999/]
- 120. Genome-wide CRISPR guide RNA design and specificity ... [https://genomebiology.biomedcentral.com/articles/10.1186/s13059-025-03488-8]
- 121. How to Pick the Best CRISPR Data Analysis Method for ... [https://www.synthego.com/guide/how-to-use-crispr/analysis-methods/]
- 122. CRISPR Gene Editing Workflow: A Step-by-Step Guide [https://www.cd-genomics.com/blog/crispr-gene-editing-workflow-a-step-by-step-guide/]
- 123. Plan Your CRISPR Experiment [https://www.addgene.org/guides/crispr/plan-your-experiment/]
- 124. Comprehensive benchmarking of genome editing ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12144422/]
- 125. Precise measurement of CRISPR genome editing ... [https://www.sciencedirect.com/science/article/pii/S2329050125000440]
- 126. Validating CRISPR/Cas9-mediated Gene Editing [https://www.sigmaaldrich.com/US/en/technical-documents/technical-article/genomics/advanced-gene-editing/validating-crispr-cas9-mediated-gene-editing?srsltid=AfmBOorrEOjT4EpxA6fWYAbY0_9koYcnfmdcErrD9oDwByS5ab77K5va]
- 127. CRISPR workflow for genome editing | IDT [https://www.idtdna.com/pages/technology/crispr/crispr-workflow]
- 128. Press Release [https://ir.crisprtx.com/news-releases/news-release-details/crispr-therapeutics-provides-business-update-and-reports-third-6]
- 129. Press Release [https://ir.crisprtx.com/news-releases/news-release-details/crispr-therapeutics-announces-positive-phase-1-clinical-data]
- 130. First-in-human trial of CRISPR gene-editing therapy safely ... [https://newsroom.heart.org/news/first-in-human-trial-of-crispr-gene-editing-therapy-safely-lowered-cholesterol-triglycerides]
- 131. Revolutionizing CRISPR technology with artificial intelligence [https://www.nature.com/articles/s12276-025-01462-9]
- 132. ZFN, TALEN and CRISPR/Cas-based methods for genome ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC3694601/]
- 133. Expanding the genetic editing tool kit: ZFNs, TALENs, and ... [https://www.jci.org/articles/view/72992]
- 134. CRISPR-Cas9, TALENs and ZFNs - the battle in gene editing [https://www.ptglab.com/news/blog/crispr-cas9-talens-and-zfns-the-battle-in-gene-editing/?srsltid=AfmBOortlMm-SbMk_DUXQxRACkdFKHBOgaz-RZrvFqjkr8ljNOJxhJcg]
- 135. The comparison of ZFNs, TALENs, and SpCas9 by GUIDE- ... [https://www.sciencedirect.com/science/article/pii/S216225312100202X]
- 136. CRISPR-Cas9 vs TALEN vs ZFN: Precision Gene Editing ... [https://synapse.patsnap.com/article/crispr-cas9-vs-talen-vs-zfn-precision-gene-editing-compared]
- 137. Genome Editing Market Report 2025: CRISPR and Beyond [https://finance.yahoo.com/news/genome-editing-market-report-2025-151500990.html]
- 138. Genome Editing Market to Grow at 16.9% CAGR [https://www.bccresearch.com/pressroom/bio/genome-editing-market-to-grow-at-169-cagr?srsltid=AfmBOoq6FWyoEjDjhLm5SwpcVhoFvdKTrDb0NgTBB6CoU6JZGH1XvyT0]
- 139. how conformational regulation of CRISPR-Cas effectors ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12599258/]
- 140. CRISPR 101: Cas9 vs. The Other Cas(s) [https://blog.addgene.org/crispr-101-cas9-vs.-the-other-cass]
- 141. Comparison of CRISPR-Cas9, CRISPR-Cas12f1, and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12131857/]
- 142. Recent advances in CRISPR-based single-nucleotide ... [https://www.nature.com/articles/s43856-025-00933-4]
- 143. CRISPR Gene Therapy: Applications, Limitations, and ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7427626/]
- 144. Past, present, and future of CRISPR genome editing ... [https://www.sciencedirect.com/science/article/pii/S0092867424001119]
- 145. Advances in RNA-based therapeutics: current breakthroughs ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12592931/]
- 146. CMN Weekly (26 September 2025) [https://crisprmedicinenews.com/news/cmn-weekly-26-september-2025-your-weekly-crispr-medicine-news/]
- 147. CMN Weekly (3 October 2025) [https://crisprmedicinenews.com/news/cmn-weekly-3-october-2025-your-weekly-crispr-medicine-news/]
- 148. Understanding FDA Cell and Gene Therapy Guidance [https://www.synthego.com/blog/fda-cell-and-gene-therapy-guidance/]
- 149. Cellular & Gene Therapy Guidances [https://www.fda.gov/vaccines-blood-biologics/biologics-guidances/cellular-gene-therapy-guidances]
- 150. Press Release [https://ir.crisprtx.com/news-releases/news-release-details/crispr-therapeutics-provides-business-update-and-reports-13]
- 151. FDA unveils new regulatory roadmap for bespoke drug ... [https://www.biopharmadive.com/news/fda-plausible-mechanism-pathway-n-of-1-crispr/805235/]
- 152. FDA officials offer roadmap for gene-editing cases like Baby KJ's [https://www.statnews.com/2025/11/12/fda-roadmap-how-to-scale-baby-kj-custom-gene-editing-treatment/]
- 153. Multidisciplinary: gene therapy [https://www.ema.europa.eu/en/human-regulatory-overview/research-development/scientific-guidelines/multidisciplinary-guidelines/multidisciplinary-gene-therapy]